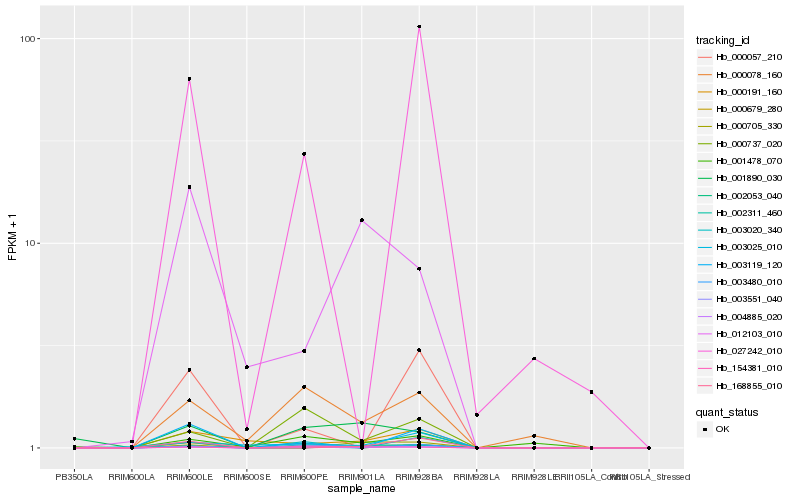
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004885_020 |
0.0 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002311_460 |
0.201468895 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000191_160 |
0.2742063899 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 4 |
Hb_000078_160 |
0.2808686961 |
- |
- |
PREDICTED: protein LURP-one-related 12-like [Jatropha curcas] |
| 5 |
Hb_154381_010 |
0.2837124816 |
- |
- |
- |
| 6 |
Hb_003551_040 |
0.2843923841 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 7 |
Hb_003025_010 |
0.3037227454 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 8 |
Hb_168855_010 |
0.3053198923 |
- |
- |
- |
| 9 |
Hb_002053_040 |
0.306466001 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 24 [Jatropha curcas] |
| 10 |
Hb_003480_010 |
0.3093457474 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 11 |
Hb_012103_010 |
0.3098836747 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 12 |
Hb_001890_030 |
0.313871046 |
- |
- |
- |
| 13 |
Hb_000737_020 |
0.313947069 |
- |
- |
- |
| 14 |
Hb_027242_010 |
0.3227801383 |
- |
- |
PREDICTED: pathogenesis-related protein 1-like [Populus euphratica] |
| 15 |
Hb_003020_340 |
0.3266302601 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 16 |
Hb_000705_330 |
0.3314005484 |
transcription factor |
TF Family: C2H2 |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 17 |
Hb_000057_210 |
0.3316466258 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 18 |
Hb_001478_070 |
0.3323472126 |
- |
- |
PREDICTED: uncharacterized protein LOC105789550 [Gossypium raimondii] |
| 19 |
Hb_003119_120 |
0.3338025332 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Nicotiana sylvestris] |
| 20 |
Hb_000679_280 |
0.3341398376 |
- |
- |
hypothetical protein POPTR_0006s27120g [Populus trichocarpa] |