| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
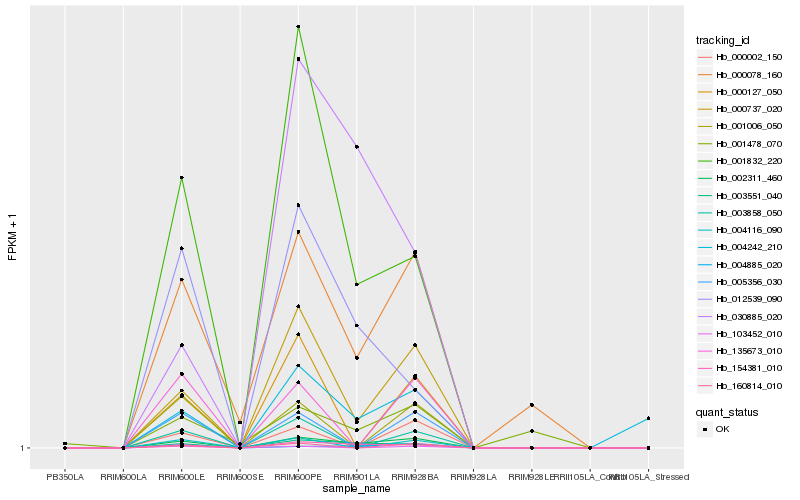
Hb_002311_460 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_003551_040 |
0.1365607818 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 3 |
Hb_000737_020 |
0.1368456226 |
- |
- |
- |
| 4 |
Hb_154381_010 |
0.1371207061 |
- |
- |
- |
| 5 |
Hb_001832_220 |
0.1774259514 |
- |
- |
- |
| 6 |
Hb_000078_160 |
0.199411733 |
- |
- |
PREDICTED: protein LURP-one-related 12-like [Jatropha curcas] |
| 7 |
Hb_004885_020 |
0.201468895 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_012539_090 |
0.257305872 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U-box domain-containing protein 26, partial [Eucalyptus grandis] |
| 9 |
Hb_004242_210 |
0.258555766 |
- |
- |
PREDICTED: uncharacterized protein LOC105633248 [Jatropha curcas] |
| 10 |
Hb_030885_020 |
0.2650703882 |
- |
- |
- |
| 11 |
Hb_160814_010 |
0.2788264562 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004116_090 |
0.282346189 |
- |
- |
- |
| 13 |
Hb_000002_150 |
0.2829599784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005356_030 |
0.2838759183 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000127_050 |
0.2866290095 |
- |
- |
- |
| 16 |
Hb_103452_010 |
0.287247516 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 17 |
Hb_001478_070 |
0.2872575372 |
- |
- |
PREDICTED: uncharacterized protein LOC105789550 [Gossypium raimondii] |
| 18 |
Hb_003858_050 |
0.2878707839 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |
| 19 |
Hb_135673_010 |
0.2910678278 |
- |
- |
Germin-like protein subfamily 1 member 6 precursor [Populus trichocarpa] |
| 20 |
Hb_001006_050 |
0.2917047807 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |