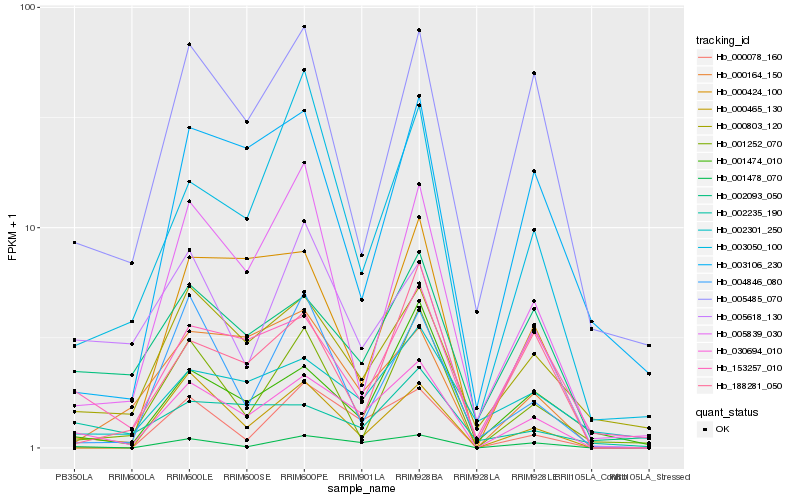
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030694_010 |
0.0 |
- |
- |
PREDICTED: endoglucanase 25-like [Solanum tuberosum] |
| 2 |
Hb_001478_070 |
0.137452403 |
- |
- |
PREDICTED: uncharacterized protein LOC105789550 [Gossypium raimondii] |
| 3 |
Hb_000803_120 |
0.1842629011 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002093_050 |
0.1915757512 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 5 |
Hb_153257_010 |
0.1928234243 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 6 |
Hb_000078_160 |
0.1979523788 |
- |
- |
PREDICTED: protein LURP-one-related 12-like [Jatropha curcas] |
| 7 |
Hb_003050_100 |
0.2008626458 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 8 |
Hb_000424_100 |
0.2014216994 |
desease resistance |
Gene Name: G-alpha |
GTP-binding protein (p) alpha subunit, gpa1, putative [Ricinus communis] |
| 9 |
Hb_001474_010 |
0.2035877129 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005839_030 |
0.2055291806 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 11 |
Hb_005485_070 |
0.2066130129 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_005618_130 |
0.2071453118 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 13 |
Hb_000164_150 |
0.2079694106 |
- |
- |
serine/threonine-specific protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000465_130 |
0.2098118803 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0001s15550g [Populus trichocarpa] |
| 15 |
Hb_002301_250 |
0.2161126734 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a [Jatropha curcas] |
| 16 |
Hb_001252_070 |
0.2174502284 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 17 |
Hb_188281_050 |
0.2182729222 |
- |
- |
PREDICTED: solute carrier family 35 member F1-like isoform X1 [Vitis vinifera] |
| 18 |
Hb_003106_230 |
0.2207504084 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 19 |
Hb_004846_080 |
0.2259766894 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 20 |
Hb_002235_190 |
0.2271655785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |