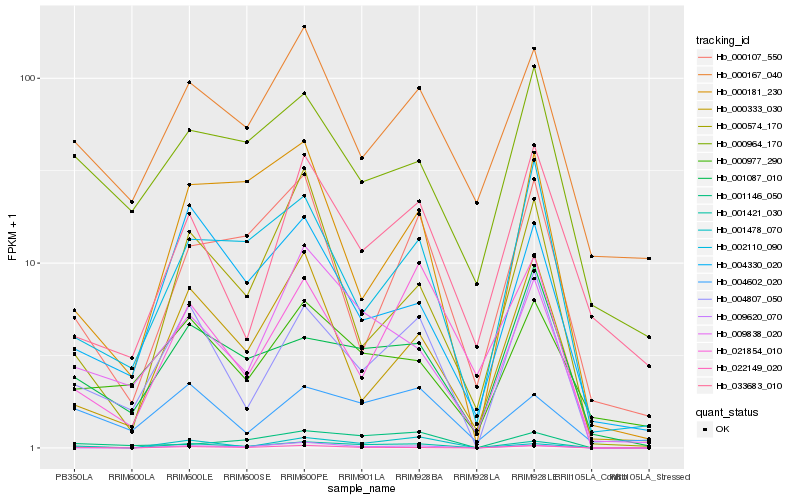
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001421_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_022149_020 |
0.1695785955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004807_050 |
0.1710184438 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 4 |
Hb_001087_010 |
0.2034314596 |
- |
- |
PREDICTED: uncharacterized protein LOC105630042 [Jatropha curcas] |
| 5 |
Hb_009838_020 |
0.2083073219 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 3 [Jatropha curcas] |
| 6 |
Hb_002110_090 |
0.2178586169 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 7 |
Hb_001146_050 |
0.2227783704 |
- |
- |
- |
| 8 |
Hb_001478_070 |
0.2268124613 |
- |
- |
PREDICTED: uncharacterized protein LOC105789550 [Gossypium raimondii] |
| 9 |
Hb_000181_230 |
0.2307344432 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_000574_170 |
0.2333893108 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 11 |
Hb_000977_290 |
0.2344787636 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_004602_020 |
0.2361299055 |
transcription factor |
TF Family: SET |
PREDICTED: N-lysine methyltransferase setd6 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004330_020 |
0.2369990773 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 14 |
Hb_021854_010 |
0.2373117135 |
- |
- |
- |
| 15 |
Hb_000333_030 |
0.2396672008 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000167_040 |
0.2397314913 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 17 |
Hb_009620_070 |
0.2400388229 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 18 |
Hb_000107_550 |
0.2426235749 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 19 |
Hb_033683_010 |
0.2442122605 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 20 |
Hb_000964_170 |
0.2447052202 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |