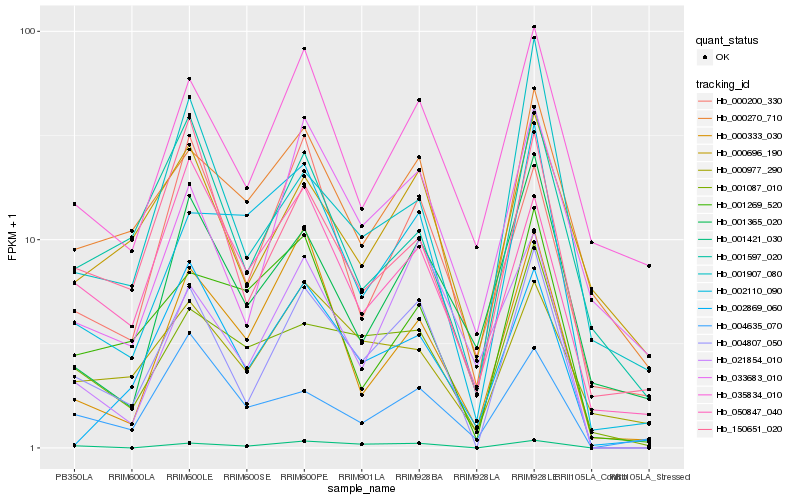
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004807_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 2 |
Hb_001421_030 |
0.1710184438 |
- |
- |
- |
| 3 |
Hb_001087_010 |
0.1883305475 |
- |
- |
PREDICTED: uncharacterized protein LOC105630042 [Jatropha curcas] |
| 4 |
Hb_021854_010 |
0.1949362048 |
- |
- |
- |
| 5 |
Hb_000270_710 |
0.1978777479 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_150651_020 |
0.1981571656 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 7 |
Hb_002110_090 |
0.1997517743 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 8 |
Hb_035834_010 |
0.202658598 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000696_190 |
0.2045886535 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 1 [Jatropha curcas] |
| 10 |
Hb_000333_030 |
0.2083186248 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 11 |
Hb_033683_010 |
0.2097930513 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 12 |
Hb_000200_330 |
0.2111743531 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_001597_020 |
0.2112810215 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001365_020 |
0.2133672877 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 15 |
Hb_002869_060 |
0.2139319491 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 16 |
Hb_000977_290 |
0.2162154048 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001907_080 |
0.2162488581 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 18 |
Hb_001269_520 |
0.2170736951 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 19 |
Hb_004635_070 |
0.2172776605 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Solanum lycopersicum] |
| 20 |
Hb_050847_040 |
0.2184947224 |
- |
- |
PREDICTED: uncharacterized protein LOC105649141 [Jatropha curcas] |