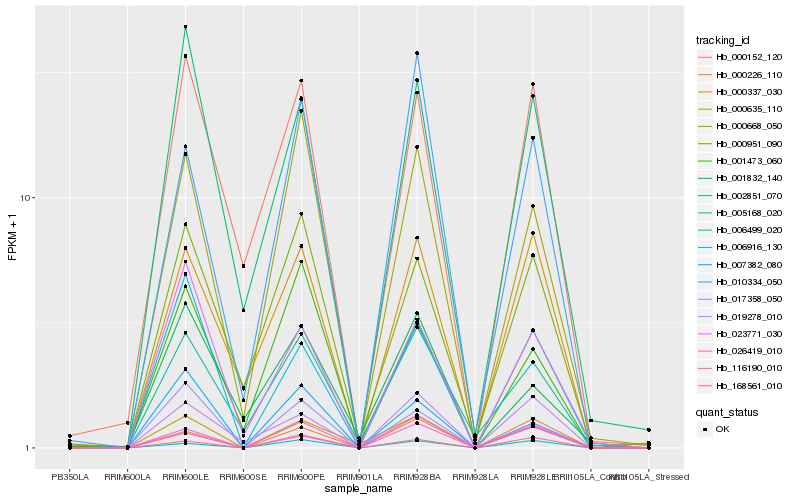
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_110 |
0.0 |
- |
- |
ELMO domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_005168_020 |
0.1597370779 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 3 |
Hb_000668_050 |
0.1741895679 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 4 |
Hb_017358_050 |
0.1785974913 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 5 |
Hb_006916_130 |
0.1810156922 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 6 |
Hb_116190_010 |
0.1848339776 |
- |
- |
PREDICTED: uncharacterized protein LOC105118535 [Populus euphratica] |
| 7 |
Hb_026419_010 |
0.1850746658 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 8 |
Hb_006499_020 |
0.1878491241 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 9 |
Hb_001832_140 |
0.1881757742 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 10 |
Hb_002851_070 |
0.1898501127 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 11 |
Hb_010334_050 |
0.1920937685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007382_080 |
0.1938535047 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 13 |
Hb_000951_090 |
0.1955559646 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 14 |
Hb_023771_030 |
0.1963245726 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000226_110 |
0.199345254 |
- |
- |
- |
| 16 |
Hb_000152_120 |
0.2005724044 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 17 |
Hb_168561_010 |
0.2017131468 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |
| 18 |
Hb_019278_010 |
0.2023458831 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 19 |
Hb_001473_060 |
0.202839953 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 20 |
Hb_000337_030 |
0.2037754711 |
- |
- |
amino acid transporter, putative [Ricinus communis] |