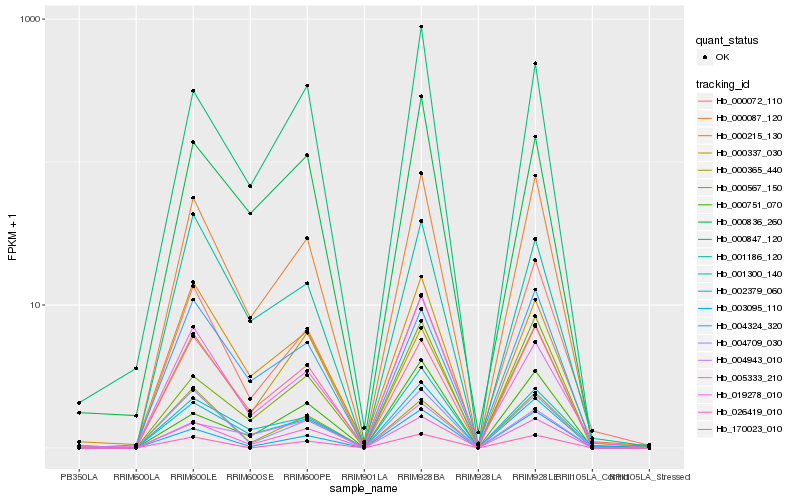
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_120 |
0.0 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 2 |
Hb_002379_060 |
0.0605682801 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 3 |
Hb_019278_010 |
0.0772868602 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 4 |
Hb_170023_010 |
0.0919767696 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 5 |
Hb_004709_030 |
0.0928306562 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 6 |
Hb_004324_320 |
0.1124357403 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 7 |
Hb_000215_130 |
0.1215712896 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 8 |
Hb_000072_110 |
0.1250154261 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 9 |
Hb_000365_440 |
0.1257499367 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000567_150 |
0.127334824 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 11 |
Hb_026419_010 |
0.1303530661 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 12 |
Hb_000847_120 |
0.1341287383 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 13 |
Hb_004943_010 |
0.1341546876 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_001186_120 |
0.1342039526 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 15 |
Hb_000836_260 |
0.1365693498 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 16 |
Hb_001300_140 |
0.1371022855 |
- |
- |
- |
| 17 |
Hb_003095_110 |
0.1399565341 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 18 |
Hb_000337_030 |
0.1476633401 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_005333_210 |
0.1489932891 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 20 |
Hb_000751_070 |
0.1491901162 |
- |
- |
- |