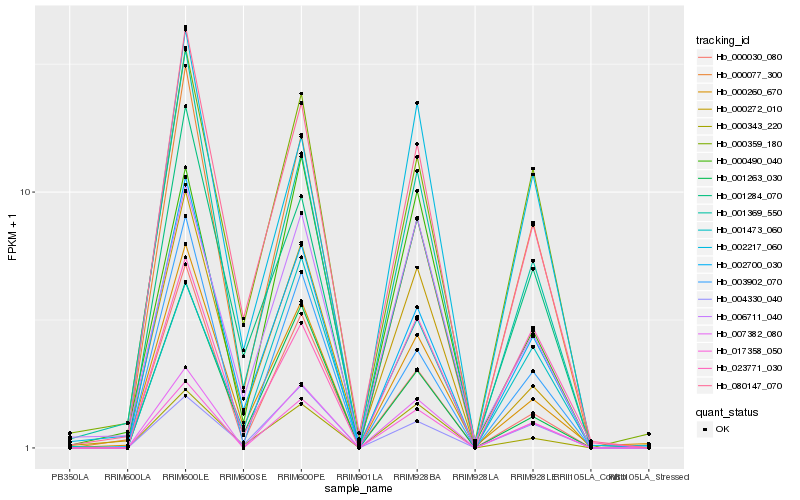
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007382_080 |
0.0 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 2 |
Hb_017358_050 |
0.0382919546 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 3 |
Hb_000343_220 |
0.1258234925 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 4 |
Hb_000490_040 |
0.1388652353 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 5 |
Hb_003902_070 |
0.1444087411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000077_300 |
0.1452170242 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 7 |
Hb_000359_180 |
0.1472921413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006711_040 |
0.1485878204 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 9 |
Hb_001473_060 |
0.149150891 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 10 |
Hb_080147_070 |
0.1505719973 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 11 |
Hb_004330_040 |
0.1516354571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000030_080 |
0.1540216679 |
- |
- |
PREDICTED: uncharacterized protein LOC105632640 [Jatropha curcas] |
| 13 |
Hb_002217_060 |
0.1548364029 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 14 |
Hb_002700_030 |
0.1561737519 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 15 |
Hb_023771_030 |
0.1562012875 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001369_550 |
0.1573865058 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 17 |
Hb_000272_010 |
0.1578715542 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 18 |
Hb_001263_030 |
0.1588195643 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 19 |
Hb_000260_670 |
0.1614763465 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 20 |
Hb_001284_070 |
0.1630179096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |