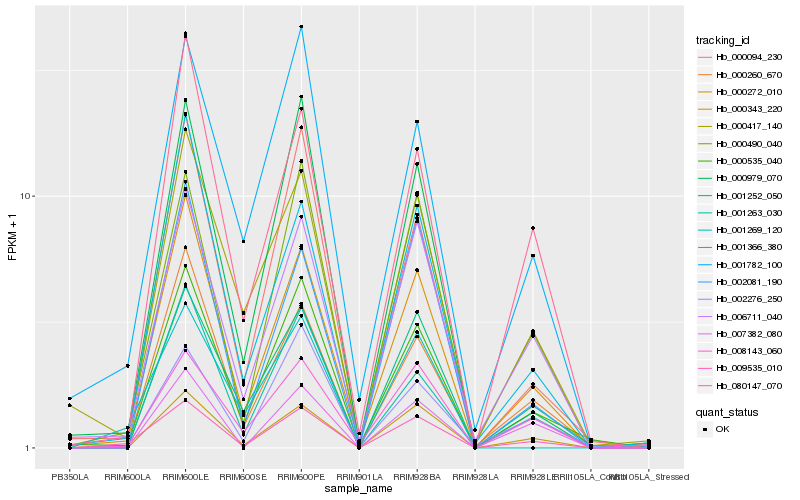
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009535_010 |
0.0 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 2 |
Hb_000490_040 |
0.1114589276 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 3 |
Hb_000272_010 |
0.1263883299 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 4 |
Hb_006711_040 |
0.1280262596 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 5 |
Hb_000343_220 |
0.1294809556 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 6 |
Hb_000535_040 |
0.1397916041 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 7 |
Hb_001252_050 |
0.1557127756 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 8 |
Hb_008143_060 |
0.1584336972 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 9 |
Hb_000094_230 |
0.1584634054 |
- |
- |
hypothetical protein JCGZ_04416 [Jatropha curcas] |
| 10 |
Hb_000260_670 |
0.1606185825 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 11 |
Hb_000979_070 |
0.1632955054 |
- |
- |
PREDICTED: endoglucanase 8-like [Jatropha curcas] |
| 12 |
Hb_001782_100 |
0.163518602 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_002081_190 |
0.1661242213 |
- |
- |
hypothetical protein EUGRSUZ_I010391, partial [Eucalyptus grandis] |
| 14 |
Hb_001263_030 |
0.1664477723 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 15 |
Hb_001366_380 |
0.1667257779 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_007382_080 |
0.1667684033 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 17 |
Hb_002276_250 |
0.1710925903 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_080147_070 |
0.1737096157 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 19 |
Hb_001269_120 |
0.1750805758 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 20 |
Hb_000417_140 |
0.1768228601 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |