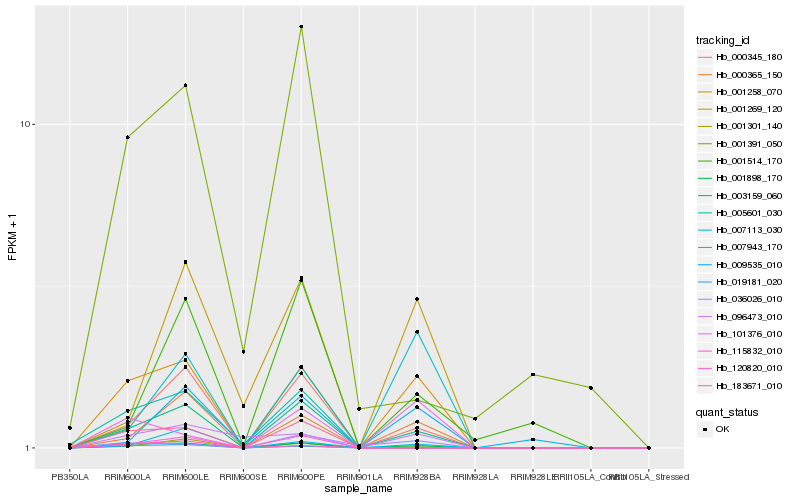
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001898_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001258_070 |
0.0979551744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003159_060 |
0.121953587 |
- |
- |
Uncharacterized protein TCM_001961 [Theobroma cacao] |
| 4 |
Hb_000345_180 |
0.2005944576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000365_150 |
0.242872258 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 6 |
Hb_001301_140 |
0.2530330852 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 7 |
Hb_183671_010 |
0.2793847976 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 8 |
Hb_036026_010 |
0.2884193206 |
transcription factor |
TF Family: FAR1 |
FAR-RED IMPAIRED RESPONSE [Medicago truncatula] |
| 9 |
Hb_101376_010 |
0.2893138889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_096473_010 |
0.2989206553 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 11 |
Hb_019181_020 |
0.3012286517 |
- |
- |
PREDICTED: uncharacterized protein LOC105763068 [Gossypium raimondii] |
| 12 |
Hb_007943_170 |
0.3061850301 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 13 |
Hb_005601_030 |
0.3079462533 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 14 |
Hb_001391_050 |
0.3089917706 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 15 |
Hb_115832_010 |
0.3126352374 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_120820_010 |
0.3272362508 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 17 |
Hb_007113_030 |
0.3278350083 |
- |
- |
- |
| 18 |
Hb_009535_010 |
0.3293411534 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 19 |
Hb_001269_120 |
0.3296608347 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 20 |
Hb_001514_170 |
0.3308116698 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |