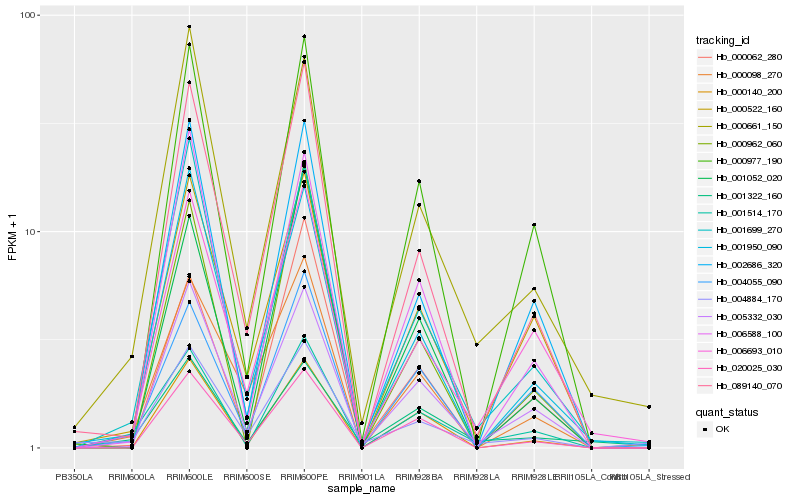
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001514_170 |
0.0 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 2 |
Hb_005332_030 |
0.1488461394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001699_270 |
0.1503127819 |
- |
- |
PREDICTED: uncharacterized protein LOC105628996 [Jatropha curcas] |
| 4 |
Hb_002686_320 |
0.1530278666 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 5 |
Hb_004055_090 |
0.1534907696 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 6 |
Hb_000522_160 |
0.1543016705 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 7 |
Hb_000977_190 |
0.1551612215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006693_010 |
0.1580800716 |
- |
- |
PREDICTED: uncharacterized protein LOC105628875 [Jatropha curcas] |
| 9 |
Hb_000661_150 |
0.1594088985 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 10 |
Hb_089140_070 |
0.1637868813 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 11 |
Hb_001322_160 |
0.1642897867 |
- |
- |
hypothetical protein JCGZ_03472 [Jatropha curcas] |
| 12 |
Hb_000140_200 |
0.1667359602 |
- |
- |
PREDICTED: uncharacterized protein LOC105642928 [Jatropha curcas] |
| 13 |
Hb_006588_100 |
0.1685421051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_020025_030 |
0.1700781219 |
transcription factor |
TF Family: PHD |
PREDICTED: protein Jade-1 [Populus euphratica] |
| 15 |
Hb_000062_280 |
0.1712039351 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 16 |
Hb_001052_020 |
0.1728336981 |
- |
- |
hypothetical protein JCGZ_26487 [Jatropha curcas] |
| 17 |
Hb_004884_170 |
0.175402922 |
- |
- |
hypothetical protein AALP_AA1G067000 [Arabis alpina] |
| 18 |
Hb_001950_090 |
0.1760710753 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 19 |
Hb_000962_060 |
0.1763247617 |
- |
- |
PREDICTED: uncharacterized protein LOC105646517 [Jatropha curcas] |
| 20 |
Hb_000098_270 |
0.1771850793 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |