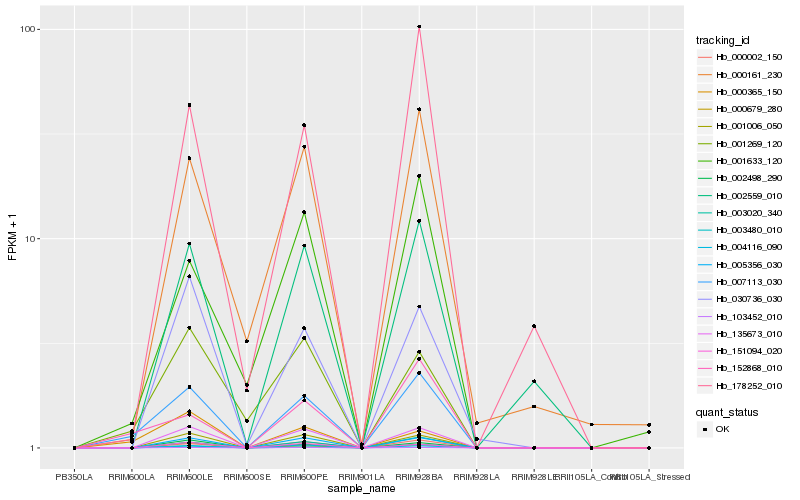
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007113_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_003020_340 |
0.1617159 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 3 |
Hb_135673_010 |
0.1685444124 |
- |
- |
Germin-like protein subfamily 1 member 6 precursor [Populus trichocarpa] |
| 4 |
Hb_152868_010 |
0.1702627675 |
rubber biosynthesis |
Gene Name: REF5 |
small rubber particle protein [Hevea brasiliensis] |
| 5 |
Hb_005356_030 |
0.1702870593 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_103452_010 |
0.1719514374 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 7 |
Hb_000002_150 |
0.172597889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001006_050 |
0.178476038 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 9 |
Hb_003480_010 |
0.192247518 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 10 |
Hb_004116_090 |
0.2002395883 |
- |
- |
- |
| 11 |
Hb_002498_290 |
0.2014124959 |
- |
- |
PREDICTED: autophagy-related protein 8C-like isoform X2 [Elaeis guineensis] |
| 12 |
Hb_000161_230 |
0.2039885518 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 13 |
Hb_002559_010 |
0.2052616595 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 14 |
Hb_001633_120 |
0.2065658937 |
- |
- |
PREDICTED: uncharacterized protein LOC105630199 [Jatropha curcas] |
| 15 |
Hb_000365_150 |
0.2070534586 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 16 |
Hb_001269_120 |
0.2077570796 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 17 |
Hb_151094_020 |
0.2082413833 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 18 |
Hb_000679_280 |
0.2084846233 |
- |
- |
hypothetical protein POPTR_0006s27120g [Populus trichocarpa] |
| 19 |
Hb_030736_030 |
0.2097352745 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 20 |
Hb_178252_010 |
0.2119491795 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |