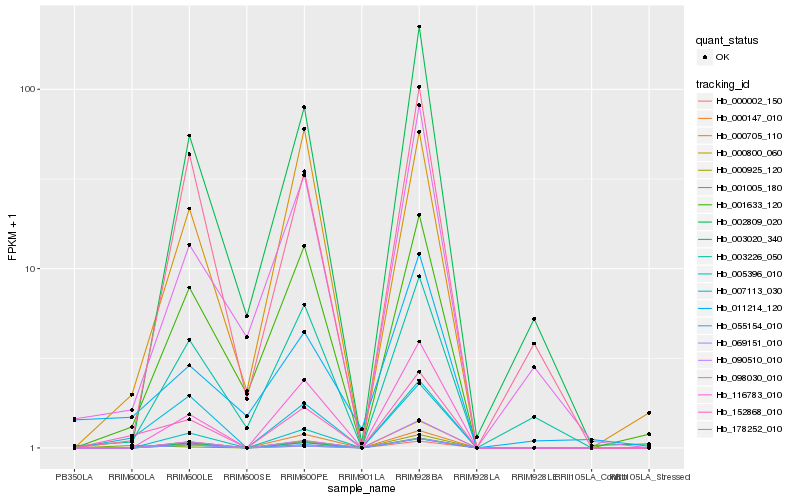
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_152868_010 |
0.0 |
rubber biosynthesis |
Gene Name: REF5 |
small rubber particle protein [Hevea brasiliensis] |
| 2 |
Hb_007113_030 |
0.1702627675 |
- |
- |
- |
| 3 |
Hb_000925_120 |
0.178102748 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 4 |
Hb_055154_010 |
0.1785796113 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 5 |
Hb_116783_010 |
0.1847143891 |
- |
- |
hypothetical protein POPTR_0011s16190g [Populus trichocarpa] |
| 6 |
Hb_001633_120 |
0.1954948263 |
- |
- |
PREDICTED: uncharacterized protein LOC105630199 [Jatropha curcas] |
| 7 |
Hb_090510_010 |
0.1985757776 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 8 |
Hb_003020_340 |
0.2044311894 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 9 |
Hb_000800_060 |
0.2062066301 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 10 |
Hb_002809_020 |
0.2065198189 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 11 |
Hb_000147_010 |
0.2072323428 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 12 |
Hb_001005_180 |
0.2112167748 |
- |
- |
hypothetical protein Csa_5G464830 [Cucumis sativus] |
| 13 |
Hb_098030_010 |
0.2136471254 |
- |
- |
hypothetical protein JCGZ_06251 [Jatropha curcas] |
| 14 |
Hb_178252_010 |
0.2146919548 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 15 |
Hb_005396_010 |
0.2163733912 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 16 |
Hb_000002_150 |
0.2197726902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011214_120 |
0.2219628015 |
- |
- |
PREDICTED: uncharacterized protein LOC105636139 [Jatropha curcas] |
| 18 |
Hb_069151_010 |
0.2231745659 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 19 |
Hb_003226_050 |
0.2255935098 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 20 |
Hb_000705_110 |
0.2279131714 |
- |
- |
hypothetical protein POPTR_0005s09220g [Populus trichocarpa] |