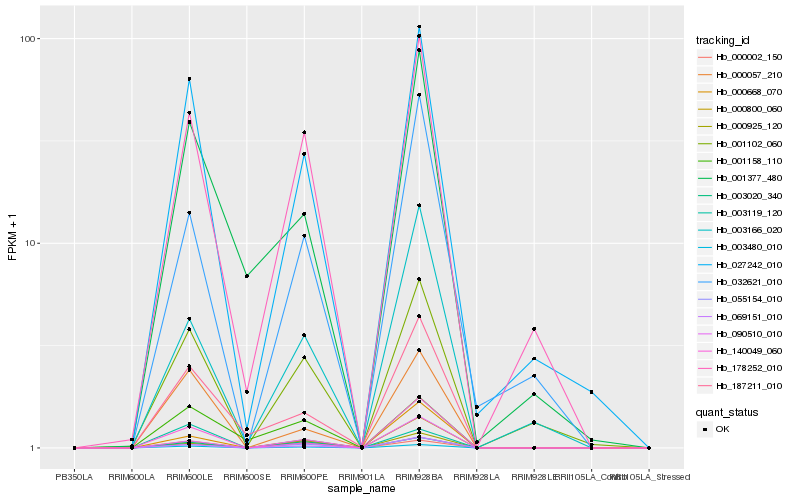
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003480_010 |
0.0 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 2 |
Hb_027242_010 |
0.0936340762 |
- |
- |
PREDICTED: pathogenesis-related protein 1-like [Populus euphratica] |
| 3 |
Hb_069151_010 |
0.113652526 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 4 |
Hb_055154_010 |
0.1170561896 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 5 |
Hb_178252_010 |
0.1214239018 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 6 |
Hb_003020_340 |
0.1266478628 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 7 |
Hb_000057_210 |
0.1282071216 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 8 |
Hb_001102_060 |
0.1411638506 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At1g78530 isoform X1 [Jatropha curcas] |
| 9 |
Hb_187211_010 |
0.1523064496 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 10 |
Hb_140049_060 |
0.1645264335 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 11 |
Hb_090510_010 |
0.1713000808 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 12 |
Hb_000800_060 |
0.1736673642 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 13 |
Hb_032621_010 |
0.1756644259 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 14 |
Hb_000925_120 |
0.1770203077 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 15 |
Hb_001377_480 |
0.1779595603 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 16 |
Hb_000002_150 |
0.1799476696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000668_070 |
0.1802795405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003166_020 |
0.1804992047 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 19 |
Hb_003119_120 |
0.1827021424 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Nicotiana sylvestris] |
| 20 |
Hb_001158_110 |
0.1832215895 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |