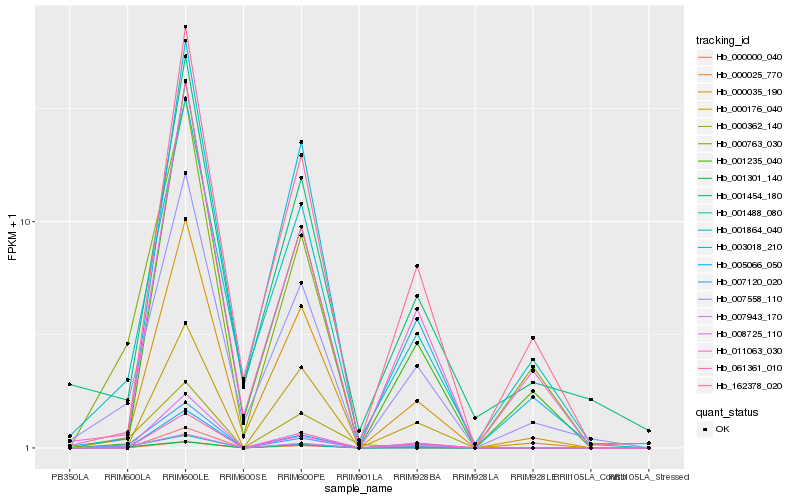
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_080 |
0.0 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_007943_170 |
0.094043158 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 3 |
Hb_001301_140 |
0.1239320185 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 4 |
Hb_007558_110 |
0.1629335163 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 5 |
Hb_000035_190 |
0.1856173062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007120_020 |
0.1873261684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005066_050 |
0.1995684472 |
- |
- |
- |
| 8 |
Hb_001864_040 |
0.2042028379 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 9 |
Hb_008725_110 |
0.2081820606 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 1 [Jatropha curcas] |
| 10 |
Hb_000025_770 |
0.2102274006 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 11 |
Hb_000000_040 |
0.2106404074 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 12 |
Hb_061361_010 |
0.2120856396 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_011063_030 |
0.2181324524 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000763_030 |
0.2181339097 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000176_040 |
0.220380007 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000362_140 |
0.2211840875 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 17 |
Hb_001235_040 |
0.2218384416 |
- |
- |
hypothetical protein POPTR_0001s47470g [Populus trichocarpa] |
| 18 |
Hb_162378_020 |
0.222364471 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 13-like [Populus euphratica] |
| 19 |
Hb_003018_210 |
0.2230721027 |
- |
- |
PREDICTED: uncharacterized protein LOC105138721 isoform X2 [Populus euphratica] |
| 20 |
Hb_001454_180 |
0.2232341108 |
- |
- |
PREDICTED: uncharacterized protein LOC105643432 [Jatropha curcas] |