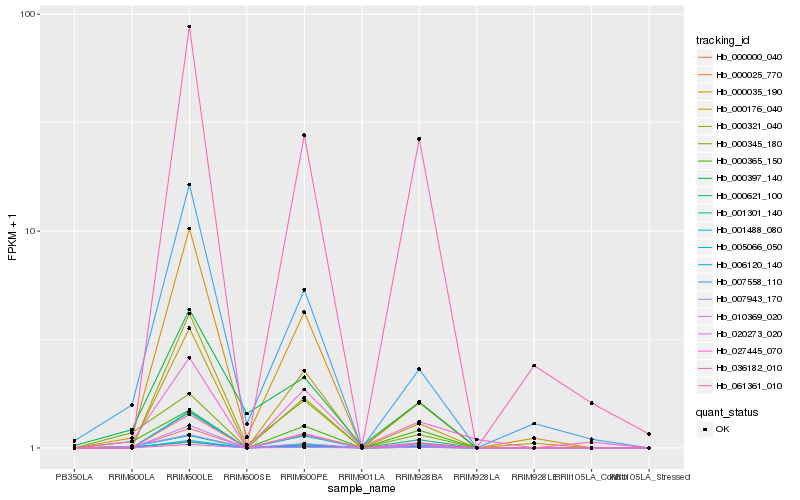
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007943_170 |
0.0 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 2 |
Hb_001488_080 |
0.094043158 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_001301_140 |
0.1182149907 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 4 |
Hb_000365_150 |
0.1382054367 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 5 |
Hb_007558_110 |
0.1736883638 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 6 |
Hb_000621_100 |
0.1826266273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006120_140 |
0.189908655 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 8 |
Hb_000025_770 |
0.1930739978 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 9 |
Hb_061361_010 |
0.1940121587 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_005066_050 |
0.1953894476 |
- |
- |
- |
| 11 |
Hb_000000_040 |
0.1969391837 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 12 |
Hb_000035_190 |
0.1978428939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000345_180 |
0.2008731077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000176_040 |
0.2030087083 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000321_040 |
0.2035432319 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 16 |
Hb_010369_020 |
0.2093651462 |
- |
- |
hypothetical protein JCGZ_21088 [Jatropha curcas] |
| 17 |
Hb_020273_020 |
0.2105514039 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_027445_070 |
0.2115121185 |
- |
- |
PREDICTED: denticleless protein homolog [Jatropha curcas] |
| 19 |
Hb_000397_140 |
0.2141607198 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 20 |
Hb_036182_010 |
0.2148690473 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |