| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
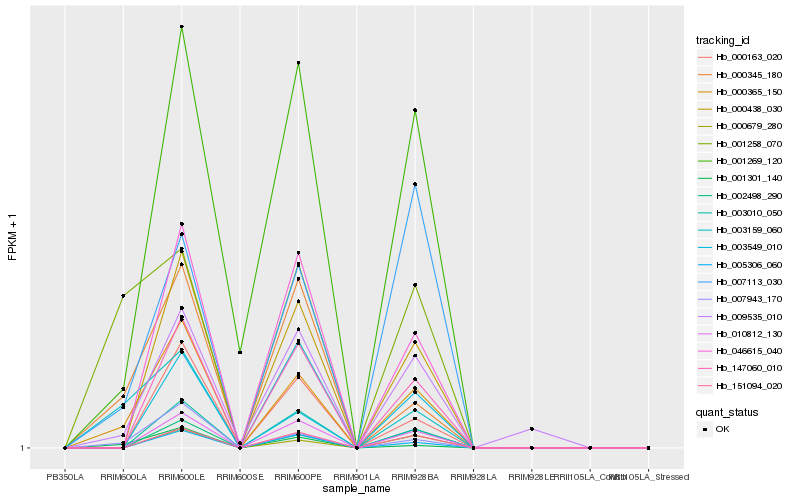
| 1 |
Hb_000365_150 |
0.0 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 2 |
Hb_007943_170 |
0.1382054367 |
- |
- |
PREDICTED: rho guanine nucleotide exchange factor 8-like [Jatropha curcas] |
| 3 |
Hb_001301_140 |
0.1720047827 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 4 |
Hb_000345_180 |
0.174197705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003159_060 |
0.1835148699 |
- |
- |
Uncharacterized protein TCM_001961 [Theobroma cacao] |
| 6 |
Hb_000438_030 |
0.1917743823 |
- |
- |
- |
| 7 |
Hb_147060_010 |
0.1974502032 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Populus euphratica] |
| 8 |
Hb_010812_130 |
0.1983090604 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 9 |
Hb_001269_120 |
0.198692002 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 10 |
Hb_009535_010 |
0.1990113951 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 11 |
Hb_003010_050 |
0.202306596 |
- |
- |
- |
| 12 |
Hb_046615_040 |
0.2048648709 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_151094_020 |
0.2050164686 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Prunus mume] |
| 14 |
Hb_007113_030 |
0.2070534586 |
- |
- |
- |
| 15 |
Hb_002498_290 |
0.2080654129 |
- |
- |
PREDICTED: autophagy-related protein 8C-like isoform X2 [Elaeis guineensis] |
| 16 |
Hb_005306_060 |
0.2087835527 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 17 |
Hb_001258_070 |
0.2092769243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000163_020 |
0.210313757 |
- |
- |
- |
| 19 |
Hb_003549_010 |
0.2118400055 |
- |
- |
PREDICTED: uncharacterized protein LOC105647434 [Jatropha curcas] |
| 20 |
Hb_000679_280 |
0.2142255468 |
- |
- |
hypothetical protein POPTR_0006s27120g [Populus trichocarpa] |