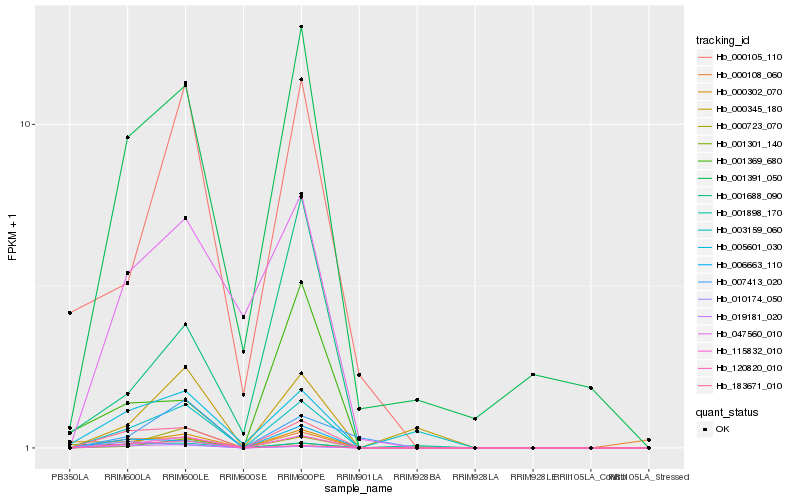
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183671_010 |
0.0 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_115832_010 |
0.1373806705 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_006663_110 |
0.1490882505 |
- |
- |
PREDICTED: U-box domain-containing protein 34-like [Camelina sativa] |
| 4 |
Hb_005601_030 |
0.1520309884 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 5 |
Hb_019181_020 |
0.1876345786 |
- |
- |
PREDICTED: uncharacterized protein LOC105763068 [Gossypium raimondii] |
| 6 |
Hb_001391_050 |
0.207531547 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 7 |
Hb_120820_010 |
0.2411670527 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 8 |
Hb_047560_010 |
0.2559715113 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 9 |
Hb_003159_060 |
0.2744632399 |
- |
- |
Uncharacterized protein TCM_001961 [Theobroma cacao] |
| 10 |
Hb_000345_180 |
0.2779102947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001898_170 |
0.2793847976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000302_070 |
0.2854011625 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 13 |
Hb_000105_110 |
0.2995463745 |
- |
- |
hypothetical protein, partial [Fervidibacteria bacterium SCGC AAA471-D06] |
| 14 |
Hb_001369_680 |
0.3023618328 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 15 |
Hb_001688_090 |
0.3027479878 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |
| 16 |
Hb_007413_020 |
0.3069618793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001301_140 |
0.3129325954 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 18 |
Hb_000108_060 |
0.3161897058 |
- |
- |
hypothetical protein POPTR_0010s07820g [Populus trichocarpa] |
| 19 |
Hb_010174_050 |
0.3193841103 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 20 |
Hb_000723_070 |
0.3244555293 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |