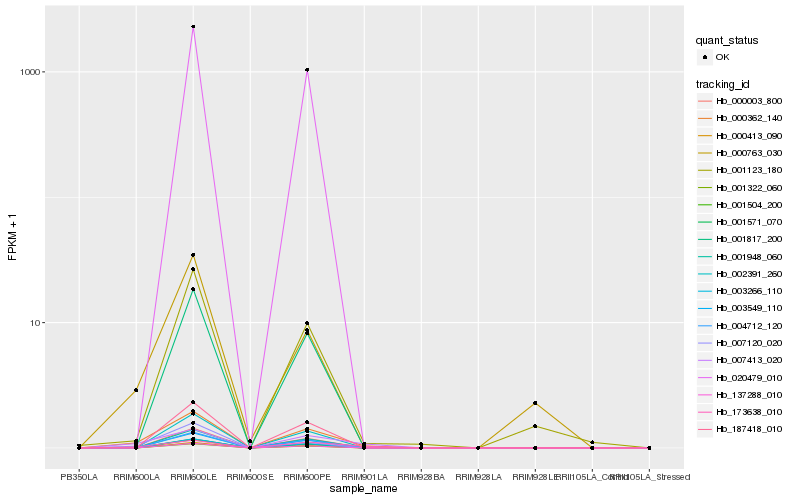
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_140 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 2 |
Hb_007413_020 |
0.1549373389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_173638_010 |
0.1707321867 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 4 |
Hb_007120_020 |
0.193218985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000763_030 |
0.2074071697 |
- |
- |
Taxadien-5-alpha-ol O-acetyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001123_180 |
0.2090303549 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_137288_010 |
0.2140334087 |
- |
- |
hypothetical protein B456_003G055000 [Gossypium raimondii] |
| 8 |
Hb_000003_800 |
0.2140341941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001322_060 |
0.2140342465 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_003266_110 |
0.2140365369 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_001948_060 |
0.2140696277 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_020479_010 |
0.2140953995 |
- |
- |
cinnamyl alcohol dehydrogenase [Pyrus pyrifolia] |
| 13 |
Hb_000413_090 |
0.2141442171 |
- |
- |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 14 |
Hb_187418_010 |
0.2141663735 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 15 |
Hb_001571_070 |
0.2142667254 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 16 |
Hb_002391_260 |
0.214306401 |
- |
- |
hypothetical protein RCOM_0017280 [Ricinus communis] |
| 17 |
Hb_001504_200 |
0.2143748998 |
- |
- |
Beta-3 adrenergic receptor, putative [Theobroma cacao] |
| 18 |
Hb_001817_200 |
0.2144068966 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 19 |
Hb_004712_120 |
0.2145153274 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 20 |
Hb_003549_110 |
0.2145279774 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |