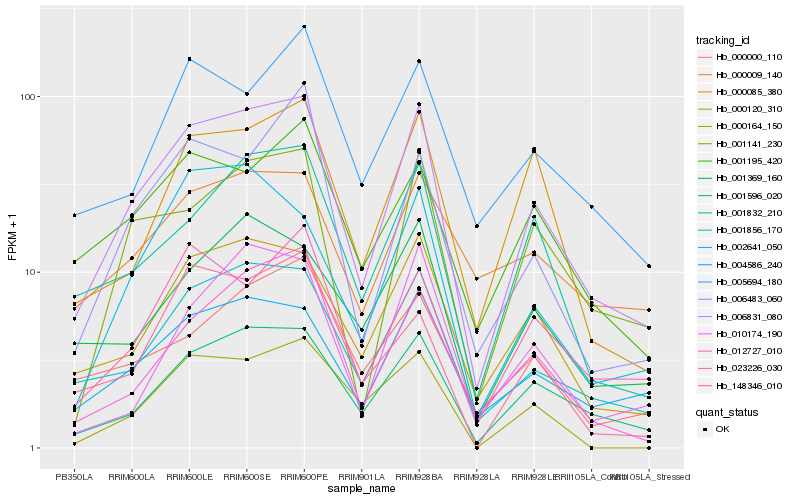
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006831_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 2 |
Hb_001596_020 |
0.1011233962 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 3 |
Hb_001856_170 |
0.1030271484 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 4 |
Hb_000120_310 |
0.1189040522 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 5 |
Hb_004586_240 |
0.1285302594 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 6 |
Hb_001141_230 |
0.1361157445 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 7 |
Hb_001832_210 |
0.1371615498 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 8 |
Hb_000085_380 |
0.1437538895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_012727_010 |
0.1438123192 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 10 |
Hb_023226_030 |
0.1460210737 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 11 |
Hb_001195_420 |
0.1528901214 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 12 |
Hb_005694_180 |
0.157913167 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 13 |
Hb_001369_160 |
0.1579725467 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 14 |
Hb_002641_050 |
0.160196882 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 15 |
Hb_006483_060 |
0.1611847907 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 16 |
Hb_010174_190 |
0.1646917708 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 17 |
Hb_000164_150 |
0.1647589635 |
- |
- |
serine/threonine-specific protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000000_110 |
0.1647611723 |
- |
- |
hypothetical protein CISIN_1g0027772mg, partial [Citrus sinensis] |
| 19 |
Hb_000009_140 |
0.1648949706 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 20 |
Hb_148346_010 |
0.1653122735 |
- |
- |
- |