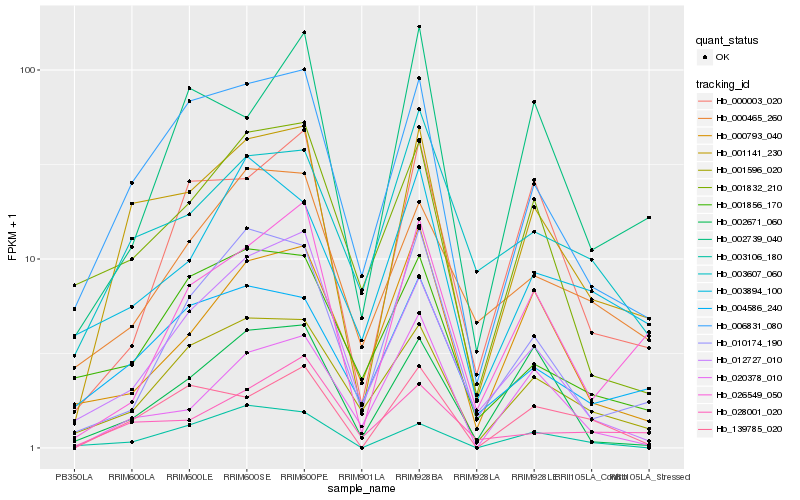
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 2 |
Hb_006831_080 |
0.1361157445 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 3 |
Hb_139785_020 |
0.1642642748 |
- |
- |
PREDICTED: uncharacterized protein LOC105641137 [Jatropha curcas] |
| 4 |
Hb_001596_020 |
0.1689713987 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 5 |
Hb_004586_240 |
0.176130735 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 6 |
Hb_000003_020 |
0.1877785333 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 7 |
Hb_028001_020 |
0.1886210358 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 8 |
Hb_001832_210 |
0.1887639371 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 9 |
Hb_026549_050 |
0.1901389644 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002671_060 |
0.1906746696 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 11 |
Hb_003607_060 |
0.1912169476 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_010174_190 |
0.1952597586 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 13 |
Hb_012727_010 |
0.1998360817 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 14 |
Hb_000465_260 |
0.2011768955 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 15 |
Hb_003106_180 |
0.2024804403 |
- |
- |
Peroxidase 65 precursor, putative [Ricinus communis] |
| 16 |
Hb_000793_040 |
0.2026744837 |
- |
- |
PREDICTED: transcription factor LHW [Jatropha curcas] |
| 17 |
Hb_003894_100 |
0.2031262724 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 18 |
Hb_001856_170 |
0.2034155576 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 19 |
Hb_002739_040 |
0.2043339538 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 20 |
Hb_020378_010 |
0.2046762748 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |