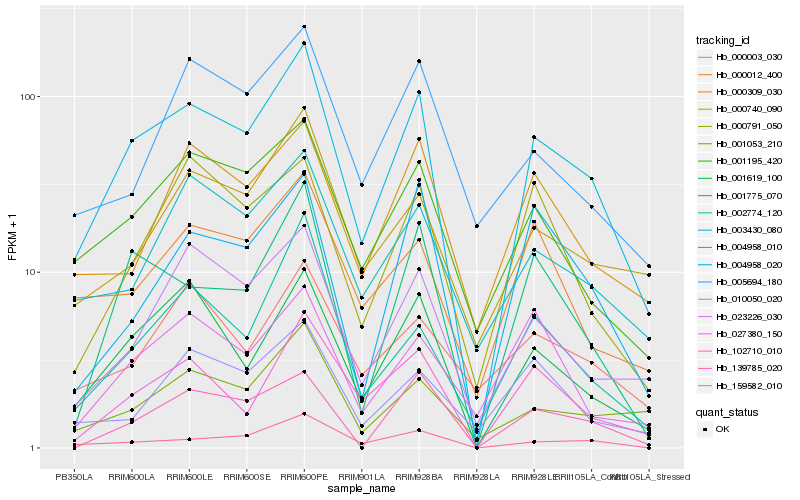
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004958_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001619_100 |
0.1325938146 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 3 |
Hb_001195_420 |
0.1450468026 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 4 |
Hb_102710_010 |
0.1518295194 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 5 |
Hb_159582_010 |
0.1529888594 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_023226_030 |
0.1554206349 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 7 |
Hb_001775_070 |
0.1579948677 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 8 |
Hb_003430_080 |
0.1631143937 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_002774_120 |
0.1652925729 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 10 |
Hb_000740_090 |
0.1660485607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001053_210 |
0.1668146939 |
- |
- |
- |
| 12 |
Hb_000309_030 |
0.1702350508 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000012_400 |
0.1729853689 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 14 |
Hb_004958_020 |
0.1758752209 |
- |
- |
PREDICTED: uncharacterized protein LOC104609039 [Nelumbo nucifera] |
| 15 |
Hb_000791_050 |
0.1782200344 |
- |
- |
PREDICTED: uncharacterized protein LOC105636760 [Jatropha curcas] |
| 16 |
Hb_139785_020 |
0.1796021654 |
- |
- |
PREDICTED: uncharacterized protein LOC105641137 [Jatropha curcas] |
| 17 |
Hb_005694_180 |
0.1804598172 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 18 |
Hb_010050_020 |
0.1817852493 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 19 |
Hb_000003_030 |
0.1829900533 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_027380_150 |
0.1850664393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |