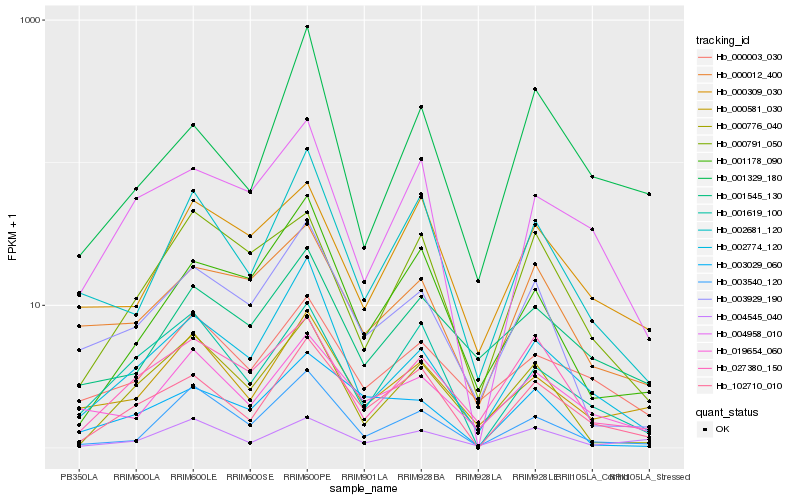
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_102710_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 2 |
Hb_004958_010 |
0.1518295194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001619_100 |
0.1533605411 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 4 |
Hb_002681_120 |
0.1612481139 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 5 |
Hb_002774_120 |
0.1853877443 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 6 |
Hb_000581_030 |
0.1872810714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_027380_150 |
0.1882780775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001178_090 |
0.1887455933 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 9 |
Hb_003029_060 |
0.1900821463 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_019654_060 |
0.1903226964 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_003540_120 |
0.1922659477 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 12 |
Hb_003929_190 |
0.1931147935 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 13 |
Hb_000309_030 |
0.1946754156 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000003_030 |
0.1950639048 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000776_040 |
0.1952948703 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X3 [Jatropha curcas] |
| 16 |
Hb_004545_040 |
0.1975973798 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2-like [Jatropha curcas] |
| 17 |
Hb_001329_180 |
0.1983170708 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 18 |
Hb_000012_400 |
0.1995164736 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 19 |
Hb_001545_130 |
0.2014575058 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 20 |
Hb_000791_050 |
0.2036836256 |
- |
- |
PREDICTED: uncharacterized protein LOC105636760 [Jatropha curcas] |