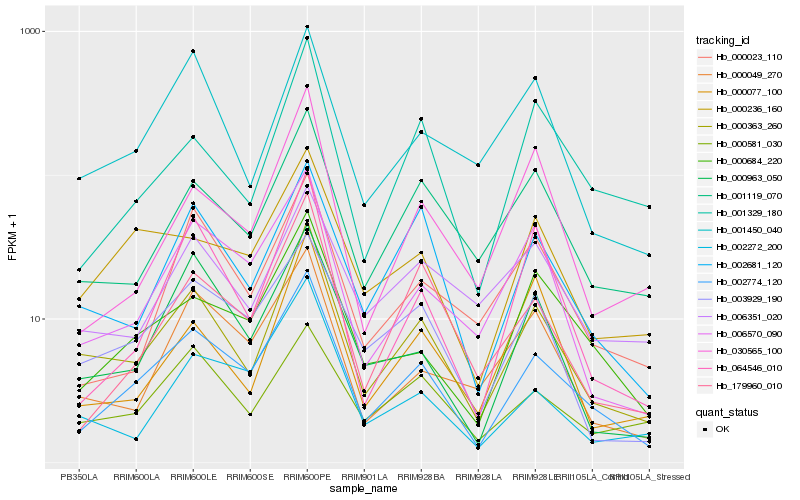
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_260 |
0.0 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 2 |
Hb_002774_120 |
0.1273847088 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 3 |
Hb_001119_070 |
0.1416748951 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 4 |
Hb_002681_120 |
0.1436585022 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 5 |
Hb_001450_040 |
0.157552291 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 6 |
Hb_003929_190 |
0.1589630199 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 7 |
Hb_000077_100 |
0.159848112 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_006570_090 |
0.1611997099 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 9 |
Hb_000963_050 |
0.1629058988 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 10 |
Hb_006351_020 |
0.1649570378 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 11 |
Hb_001329_180 |
0.1692314795 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 12 |
Hb_000049_270 |
0.1693462835 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000581_030 |
0.1699885104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002272_200 |
0.1699949641 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 15 |
Hb_064546_010 |
0.1712500492 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 16 |
Hb_179960_010 |
0.1716255981 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 17 |
Hb_000236_160 |
0.1730119025 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 18 |
Hb_030565_100 |
0.1741041802 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000023_110 |
0.1776562691 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 20 |
Hb_000684_220 |
0.1787946395 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |