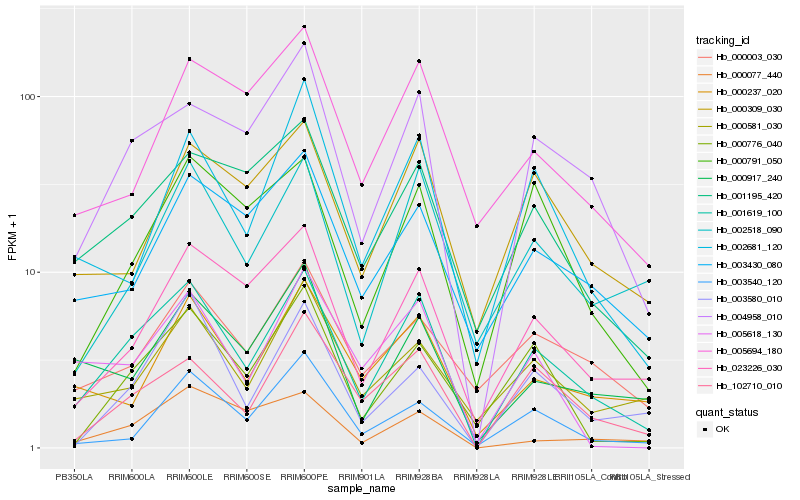
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 2 |
Hb_004958_010 |
0.1325938146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_102710_010 |
0.1533605411 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 4 |
Hb_023226_030 |
0.1595617225 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 5 |
Hb_001195_420 |
0.1665607764 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 6 |
Hb_002518_090 |
0.1717521562 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 7 |
Hb_000791_050 |
0.1775376921 |
- |
- |
PREDICTED: uncharacterized protein LOC105636760 [Jatropha curcas] |
| 8 |
Hb_002681_120 |
0.1786742581 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 9 |
Hb_000581_030 |
0.1791653243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000237_020 |
0.1810940376 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 11 |
Hb_000917_240 |
0.1811025858 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 12 |
Hb_000776_040 |
0.181902494 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X3 [Jatropha curcas] |
| 13 |
Hb_003430_080 |
0.1827015336 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000003_030 |
0.1836013866 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_000309_030 |
0.1838798309 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_005618_130 |
0.1841526513 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 17 |
Hb_003580_010 |
0.1898771543 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 18 |
Hb_005694_180 |
0.1908664868 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 19 |
Hb_000077_440 |
0.1915815436 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 20 |
Hb_003540_120 |
0.1948647746 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |