| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
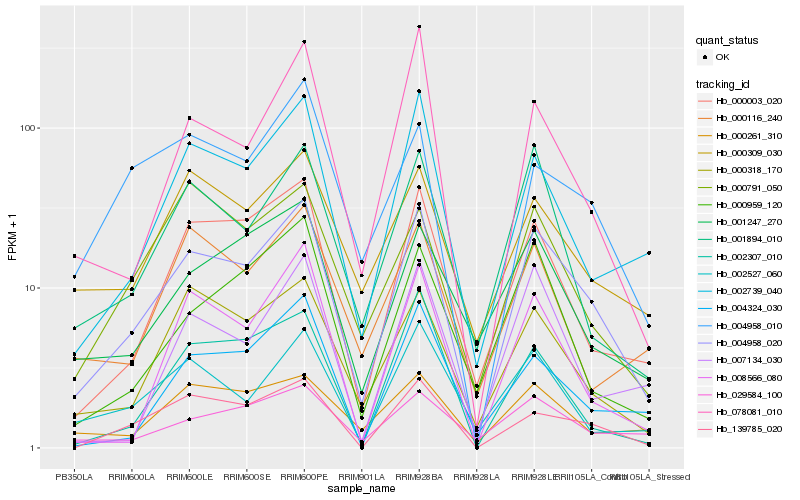
Hb_004958_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104609039 [Nelumbo nucifera] |
| 2 |
Hb_000003_020 |
0.1388300414 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 3 |
Hb_002739_040 |
0.1408692333 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 4 |
Hb_000318_170 |
0.1499166617 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 5 |
Hb_139785_020 |
0.1505282556 |
- |
- |
PREDICTED: uncharacterized protein LOC105641137 [Jatropha curcas] |
| 6 |
Hb_001894_010 |
0.1512318489 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 7 |
Hb_078081_010 |
0.1612657761 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 8 |
Hb_029584_100 |
0.1621508664 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 9 |
Hb_002527_060 |
0.1653242228 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 10 |
Hb_000309_030 |
0.1683436507 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_008566_080 |
0.1686597529 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 12 |
Hb_001247_270 |
0.1698325652 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004324_030 |
0.1715450949 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 14 |
Hb_002307_010 |
0.1716924523 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 15 |
Hb_000959_120 |
0.1718358421 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 16 |
Hb_004958_010 |
0.1758752209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000261_310 |
0.1764366953 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 18 |
Hb_000791_050 |
0.1778369375 |
- |
- |
PREDICTED: uncharacterized protein LOC105636760 [Jatropha curcas] |
| 19 |
Hb_007134_030 |
0.1796134936 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 20 |
Hb_000116_240 |
0.1798766537 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |