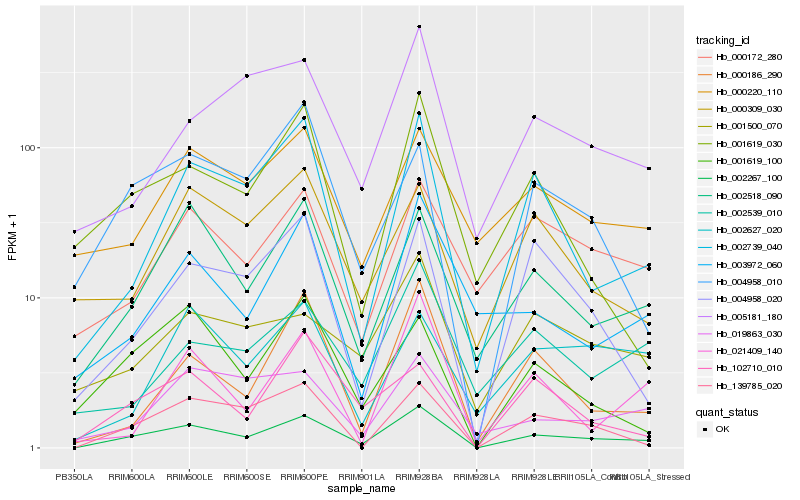
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002267_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635033 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002518_090 |
0.1778033862 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 3 |
Hb_002739_040 |
0.1823027512 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 4 |
Hb_139785_020 |
0.1823329051 |
- |
- |
PREDICTED: uncharacterized protein LOC105641137 [Jatropha curcas] |
| 5 |
Hb_000186_290 |
0.2040623892 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004958_020 |
0.2069472412 |
- |
- |
PREDICTED: uncharacterized protein LOC104609039 [Nelumbo nucifera] |
| 7 |
Hb_000172_280 |
0.2102580997 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_004958_010 |
0.2107157768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001619_100 |
0.211648103 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 10 |
Hb_001500_070 |
0.2124962855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000220_110 |
0.213474679 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 12 |
Hb_003972_060 |
0.2142811518 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 13 |
Hb_102710_010 |
0.2149488985 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 14 |
Hb_005181_180 |
0.2150578698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_019863_030 |
0.2158845022 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 16 |
Hb_002539_010 |
0.2205719672 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 17 |
Hb_001619_030 |
0.2228787245 |
- |
- |
annexin, putative [Ricinus communis] |
| 18 |
Hb_002627_020 |
0.2231409121 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 19 |
Hb_021409_140 |
0.2236463461 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 20 |
Hb_000309_030 |
0.2245774788 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |