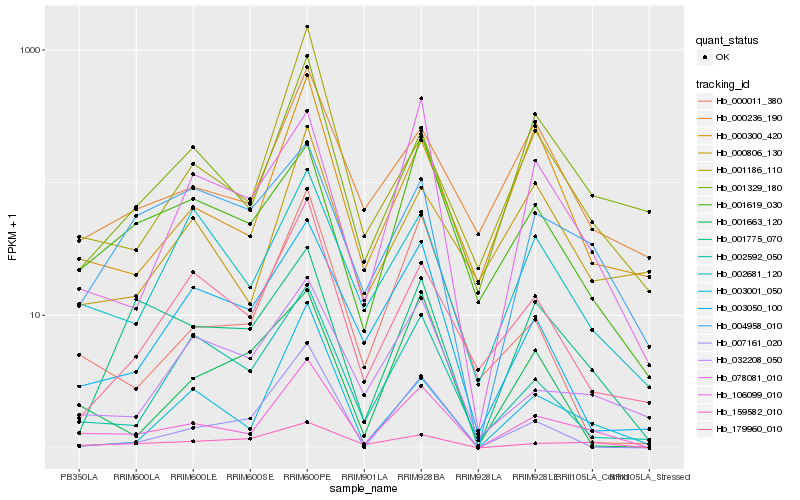
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106099_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_159582_010 |
0.1987372263 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_078081_010 |
0.2055093352 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 4 |
Hb_002681_120 |
0.2089791181 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 5 |
Hb_000300_420 |
0.2107091676 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000236_190 |
0.212190924 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 7 |
Hb_032208_050 |
0.2122782891 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 8 |
Hb_003050_100 |
0.2130840829 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 9 |
Hb_000011_380 |
0.2138501958 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 10 |
Hb_001329_180 |
0.2150359917 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 11 |
Hb_002592_050 |
0.2154025044 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 12 |
Hb_001619_030 |
0.2176349262 |
- |
- |
annexin, putative [Ricinus communis] |
| 13 |
Hb_007161_020 |
0.2180066741 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 14 |
Hb_179960_010 |
0.2205430869 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 15 |
Hb_003001_050 |
0.222363168 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 16 |
Hb_001775_070 |
0.226386871 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 17 |
Hb_001186_110 |
0.2286613136 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 18 |
Hb_004958_010 |
0.2302094512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001663_120 |
0.2316888759 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000806_130 |
0.2317218307 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |