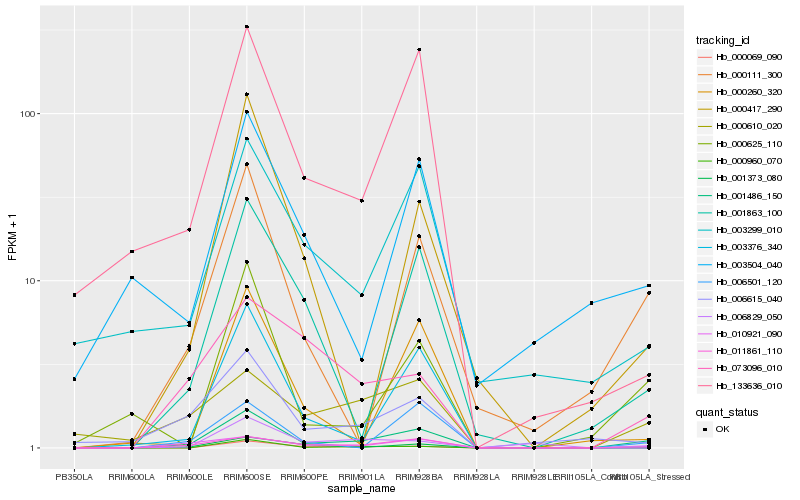
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_000960_070 |
0.1975297612 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 3 |
Hb_010921_090 |
0.2196876277 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 4 |
Hb_000111_300 |
0.2332190931 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 5 |
Hb_000069_090 |
0.2373728203 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 6 |
Hb_001863_100 |
0.250533159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_133636_010 |
0.2520390266 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 8 |
Hb_000625_110 |
0.255192224 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003299_010 |
0.258716578 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 10 |
Hb_011861_110 |
0.2597732312 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 11 |
Hb_006615_040 |
0.2607563219 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 12 |
Hb_000610_020 |
0.2625727904 |
- |
- |
unknown [Glycine max] |
| 13 |
Hb_006501_120 |
0.2637993906 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 14 |
Hb_006829_050 |
0.2672306939 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_003376_340 |
0.2711132633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001373_080 |
0.2745851086 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000417_290 |
0.2837840948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_073096_010 |
0.2846446365 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 45-like [Jatropha curcas] |
| 19 |
Hb_000260_320 |
0.2858579698 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 20 |
Hb_003504_040 |
0.2881423142 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |