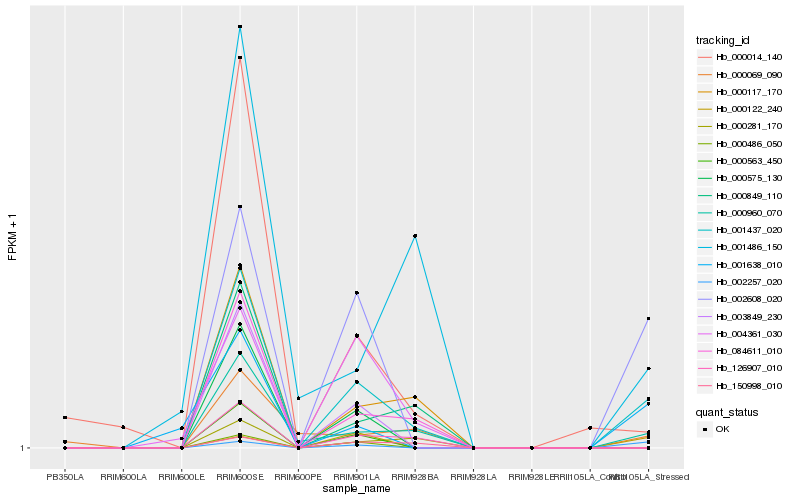
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628060 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000960_070 |
0.2093948309 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 3 |
Hb_002608_020 |
0.2171945991 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 305 [Jatropha curcas] |
| 4 |
Hb_002257_020 |
0.2805708528 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 5 |
Hb_126907_010 |
0.3005961331 |
- |
- |
- |
| 6 |
Hb_084611_010 |
0.3015690674 |
- |
- |
PREDICTED: uncharacterized protein LOC104249812 [Nicotiana sylvestris] |
| 7 |
Hb_000014_140 |
0.3017200706 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 8 |
Hb_001638_010 |
0.3035864598 |
- |
- |
- |
| 9 |
Hb_000117_170 |
0.3195365895 |
- |
- |
PREDICTED: uncharacterized protein LOC104088736 isoform X2 [Nicotiana tomentosiformis] |
| 10 |
Hb_000122_240 |
0.3203076074 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 11 |
Hb_000069_090 |
0.3243769993 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 12 |
Hb_001486_150 |
0.3329058723 |
- |
- |
- |
| 13 |
Hb_000849_110 |
0.3339484463 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 14 |
Hb_000563_450 |
0.3364321083 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 15 |
Hb_000575_130 |
0.3366209163 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Populus euphratica] |
| 16 |
Hb_003849_230 |
0.336678642 |
- |
- |
PREDICTED: NEP1-interacting protein 2-like [Jatropha curcas] |
| 17 |
Hb_000486_050 |
0.3384497036 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_150998_010 |
0.3392876502 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 19 |
Hb_000281_170 |
0.3397415337 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 20 |
Hb_004361_030 |
0.3400551201 |
- |
- |
PREDICTED: uncharacterized protein LOC104239397 isoform X1 [Nicotiana sylvestris] |