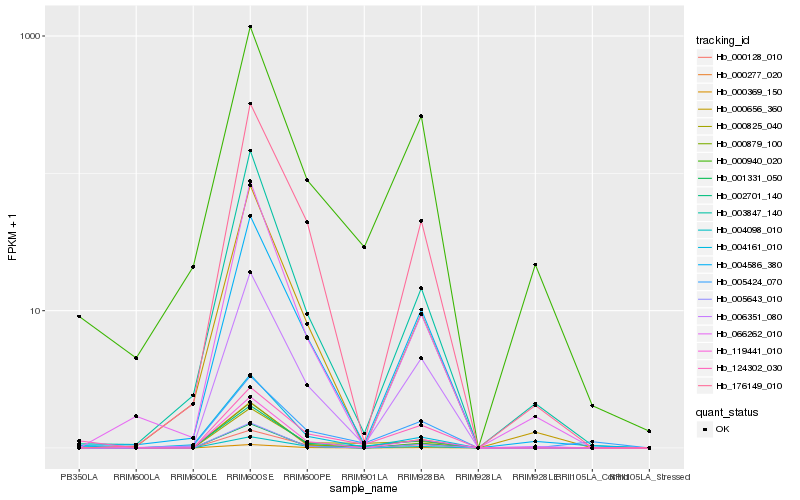
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000825_040 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_002701_140 |
0.1360744056 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 3 |
Hb_000277_020 |
0.1547586934 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 4 |
Hb_006351_080 |
0.179133056 |
- |
- |
hypothetical protein B456_011G275400 [Gossypium raimondii] |
| 5 |
Hb_119441_010 |
0.1829791959 |
- |
- |
hypothetical protein CISIN_1g0477971mg, partial [Citrus sinensis] |
| 6 |
Hb_000369_150 |
0.183725714 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20185 [Jatropha curcas] |
| 7 |
Hb_000940_020 |
0.1868328667 |
- |
- |
hypothetical protein POPTR_0004s17500g [Populus trichocarpa] |
| 8 |
Hb_004098_010 |
0.1879135897 |
- |
- |
- |
| 9 |
Hb_176149_010 |
0.1902737618 |
- |
- |
hypothetical protein JCGZ_16793 [Jatropha curcas] |
| 10 |
Hb_000128_010 |
0.1903781713 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_004586_380 |
0.191177397 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 12 |
Hb_001331_050 |
0.191297319 |
- |
- |
hypothetical protein VITISV_022439 [Vitis vinifera] |
| 13 |
Hb_005643_010 |
0.1925517673 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_005424_070 |
0.1946982003 |
- |
- |
PREDICTED: TATA-box-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_004161_010 |
0.1973983174 |
- |
- |
hypothetical protein JCGZ_21278 [Jatropha curcas] |
| 16 |
Hb_000879_100 |
0.1982311538 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 17 |
Hb_003847_140 |
0.1983364337 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000656_360 |
0.1999973679 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 19 |
Hb_066262_010 |
0.2080558791 |
- |
- |
hypothetical protein JCGZ_14798 [Jatropha curcas] |
| 20 |
Hb_124302_030 |
0.2093685877 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Beta vulgaris subsp. vulgaris] |