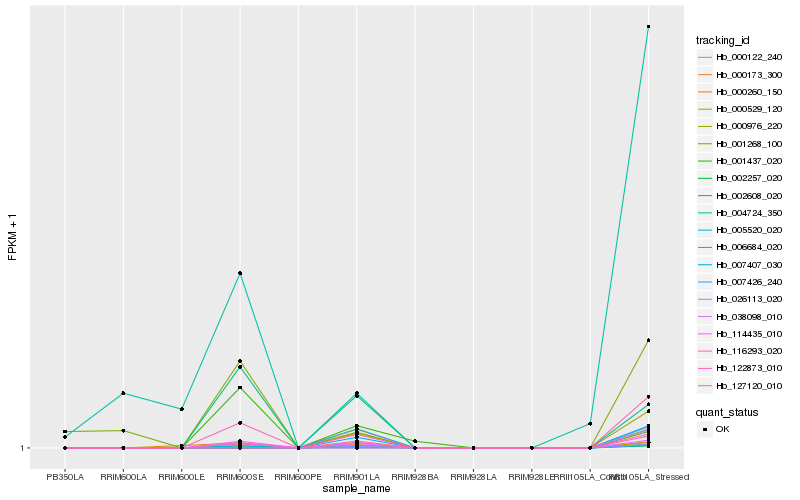
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114435_010 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_122873_010 |
0.1381140165 |
- |
- |
- |
| 3 |
Hb_038098_010 |
0.1419031404 |
- |
- |
PREDICTED: uncharacterized protein LOC105793140 [Gossypium raimondii] |
| 4 |
Hb_002257_020 |
0.1509653127 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 5 |
Hb_000122_240 |
0.1719513788 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 6 |
Hb_002608_020 |
0.2261220108 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 305 [Jatropha curcas] |
| 7 |
Hb_000260_150 |
0.305580779 |
- |
- |
hypothetical protein POPTR_0344s002001g, partial [Populus trichocarpa] |
| 8 |
Hb_001268_100 |
0.3481349168 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 9 |
Hb_004724_350 |
0.372088413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001437_020 |
0.3790779982 |
- |
- |
PREDICTED: uncharacterized protein LOC105628060 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000173_300 |
0.3802723045 |
- |
- |
PREDICTED: uncharacterized protein LOC105631894 [Jatropha curcas] |
| 12 |
Hb_026113_020 |
0.3805976179 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_006684_020 |
0.3807420253 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_000976_220 |
0.3814139717 |
- |
- |
PREDICTED: protein ALTERED XYLOGLUCAN 4-like [Jatropha curcas] |
| 15 |
Hb_127120_010 |
0.3814179092 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 16 |
Hb_007407_030 |
0.3814325138 |
- |
- |
hypothetical protein JCGZ_23510 [Jatropha curcas] |
| 17 |
Hb_000529_120 |
0.3844174651 |
- |
- |
PREDICTED: uncharacterized protein LOC105641745 [Jatropha curcas] |
| 18 |
Hb_116293_020 |
0.3876707676 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 19 |
Hb_005520_020 |
0.3876707707 |
- |
- |
hypothetical protein EUGRSUZ_G03336 [Eucalyptus grandis] |
| 20 |
Hb_007426_240 |
0.3879268119 |
- |
- |
- |