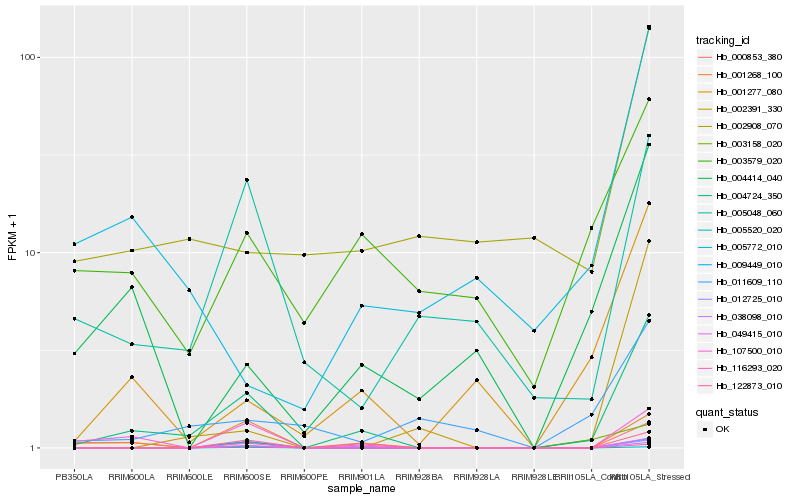
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_350 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012725_010 |
0.268816131 |
- |
- |
- |
| 3 |
Hb_122873_010 |
0.285054653 |
- |
- |
- |
| 4 |
Hb_107500_010 |
0.2863220345 |
- |
- |
- |
| 5 |
Hb_001268_100 |
0.2902203871 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 6 |
Hb_038098_010 |
0.2920170468 |
- |
- |
PREDICTED: uncharacterized protein LOC105793140 [Gossypium raimondii] |
| 7 |
Hb_049415_010 |
0.2920690421 |
- |
- |
- |
| 8 |
Hb_001277_080 |
0.3052968947 |
- |
- |
PREDICTED: vegetative cell wall protein gp1 [Jatropha curcas] |
| 9 |
Hb_005520_020 |
0.3064512861 |
- |
- |
hypothetical protein EUGRSUZ_G03336 [Eucalyptus grandis] |
| 10 |
Hb_116293_020 |
0.3069696617 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 11 |
Hb_000853_380 |
0.3149131892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005048_060 |
0.3258555964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009449_010 |
0.3302476949 |
- |
- |
Serine/threonine-protein kinase HT1 [Gossypium arboreum] |
| 14 |
Hb_004414_040 |
0.3384196965 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 15 |
Hb_002391_330 |
0.347290152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005772_010 |
0.3544434191 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 17 |
Hb_003579_020 |
0.3567117674 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 18 |
Hb_003158_020 |
0.3574452841 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 19 |
Hb_011609_110 |
0.3598865364 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 20 |
Hb_002908_070 |
0.3606370651 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |