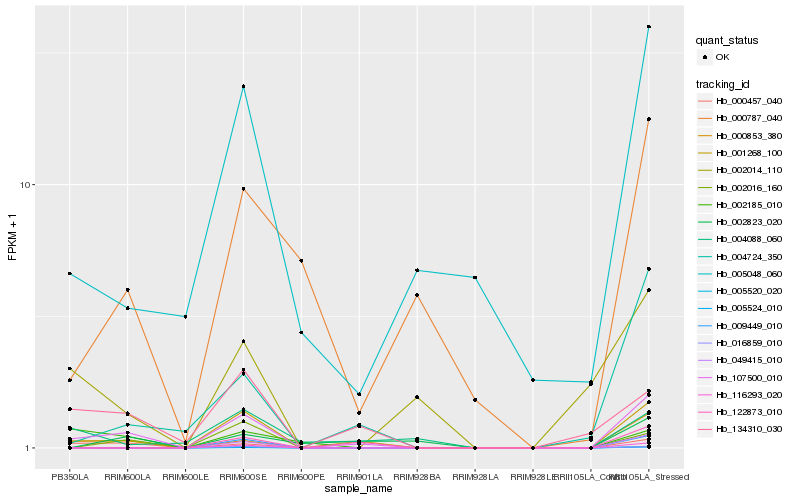
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107500_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001268_100 |
0.1137710846 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 3 |
Hb_000853_380 |
0.2261002043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000457_040 |
0.2510046801 |
- |
- |
- |
| 5 |
Hb_004724_350 |
0.2863220345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_134310_030 |
0.2956118953 |
- |
- |
hypothetical protein JCGZ_10790 [Jatropha curcas] |
| 7 |
Hb_002016_160 |
0.3160375091 |
- |
- |
- |
| 8 |
Hb_005048_060 |
0.3264734435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_116293_020 |
0.3446792458 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 10 |
Hb_005520_020 |
0.3447064092 |
- |
- |
hypothetical protein EUGRSUZ_G03336 [Eucalyptus grandis] |
| 11 |
Hb_009449_010 |
0.3462868788 |
- |
- |
Serine/threonine-protein kinase HT1 [Gossypium arboreum] |
| 12 |
Hb_049415_010 |
0.3470529341 |
- |
- |
- |
| 13 |
Hb_000787_040 |
0.3479443951 |
- |
- |
PREDICTED: uncharacterized protein LOC105115900 isoform X1 [Populus euphratica] |
| 14 |
Hb_122873_010 |
0.3543802129 |
- |
- |
- |
| 15 |
Hb_005524_010 |
0.3556493909 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 16 |
Hb_002823_020 |
0.3582273276 |
- |
- |
- |
| 17 |
Hb_004088_060 |
0.3626521586 |
- |
- |
hypothetical protein POPTR_0012s01875g [Populus trichocarpa] |
| 18 |
Hb_002014_110 |
0.3647151438 |
- |
- |
- |
| 19 |
Hb_002185_010 |
0.3665138955 |
- |
- |
PREDICTED: uncharacterized protein LOC105795658 [Gossypium raimondii] |
| 20 |
Hb_016859_010 |
0.3670074088 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |