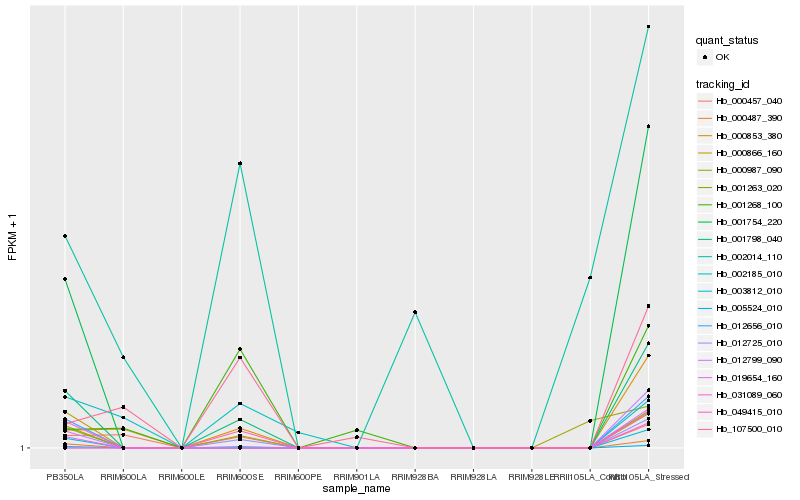
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001798_040 |
0.0 |
transcription factor |
TF Family: NAC |
hypothetical protein ARALYDRAFT_918808 [Arabidopsis lyrata subsp. lyrata] |
| 2 |
Hb_005524_010 |
0.1021495969 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 3 |
Hb_003812_010 |
0.2687831185 |
- |
- |
- |
| 4 |
Hb_012656_010 |
0.2691097052 |
- |
- |
- |
| 5 |
Hb_001263_020 |
0.2691729187 |
- |
- |
- |
| 6 |
Hb_000487_390 |
0.269477385 |
- |
- |
PREDICTED: putative pumilio homolog 21 [Jatropha curcas] |
| 7 |
Hb_001754_220 |
0.2726875218 |
- |
- |
- |
| 8 |
Hb_019654_160 |
0.2900558337 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 9 |
Hb_012799_090 |
0.2905822507 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 10 |
Hb_000853_380 |
0.3010495118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000866_160 |
0.3067225103 |
- |
- |
PREDICTED: elicitor-responsive protein 3 [Populus euphratica] |
| 12 |
Hb_031089_060 |
0.3074538526 |
- |
- |
- |
| 13 |
Hb_000457_040 |
0.3578890629 |
- |
- |
- |
| 14 |
Hb_000987_090 |
0.381435518 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 15 |
Hb_002014_110 |
0.387078546 |
- |
- |
- |
| 16 |
Hb_107500_010 |
0.3871383293 |
- |
- |
- |
| 17 |
Hb_001268_100 |
0.4010094119 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 18 |
Hb_012725_010 |
0.4037690944 |
- |
- |
- |
| 19 |
Hb_002185_010 |
0.4163118307 |
- |
- |
PREDICTED: uncharacterized protein LOC105795658 [Gossypium raimondii] |
| 20 |
Hb_049415_010 |
0.4181751317 |
- |
- |
- |