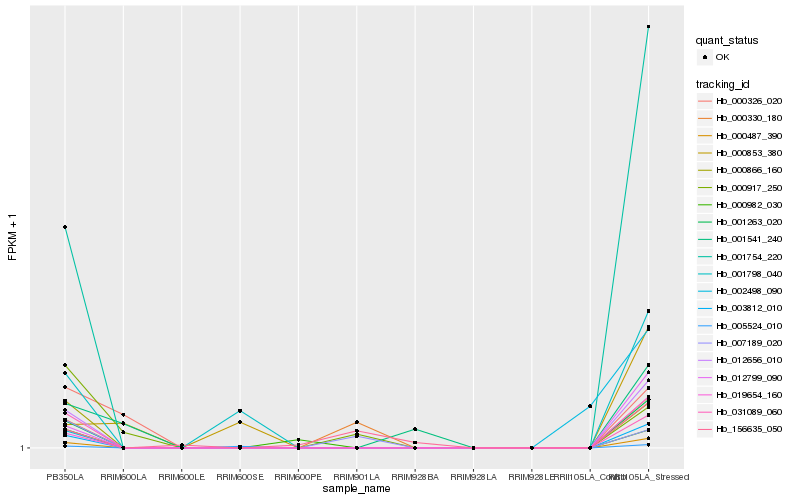
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003812_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_012656_010 |
0.0097146949 |
- |
- |
- |
| 3 |
Hb_001263_020 |
0.0109216366 |
- |
- |
- |
| 4 |
Hb_000487_390 |
0.0157738129 |
- |
- |
PREDICTED: putative pumilio homolog 21 [Jatropha curcas] |
| 5 |
Hb_001754_220 |
0.0528958163 |
- |
- |
- |
| 6 |
Hb_019654_160 |
0.1180739925 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 7 |
Hb_012799_090 |
0.1195132334 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 8 |
Hb_000866_160 |
0.1488418955 |
- |
- |
PREDICTED: elicitor-responsive protein 3 [Populus euphratica] |
| 9 |
Hb_031089_060 |
0.1504138055 |
- |
- |
- |
| 10 |
Hb_001798_040 |
0.2687831185 |
transcription factor |
TF Family: NAC |
hypothetical protein ARALYDRAFT_918808 [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_000982_030 |
0.3407658395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005524_010 |
0.3533340482 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 13 |
Hb_000326_020 |
0.3626190024 |
- |
- |
- |
| 14 |
Hb_000330_180 |
0.3634063542 |
- |
- |
- |
| 15 |
Hb_001541_240 |
0.3641087873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007189_020 |
0.3937359147 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 17 |
Hb_000853_380 |
0.4054755301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002498_090 |
0.4088480631 |
- |
- |
- |
| 19 |
Hb_156635_050 |
0.4139536712 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 20 |
Hb_000917_250 |
0.4172031939 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |