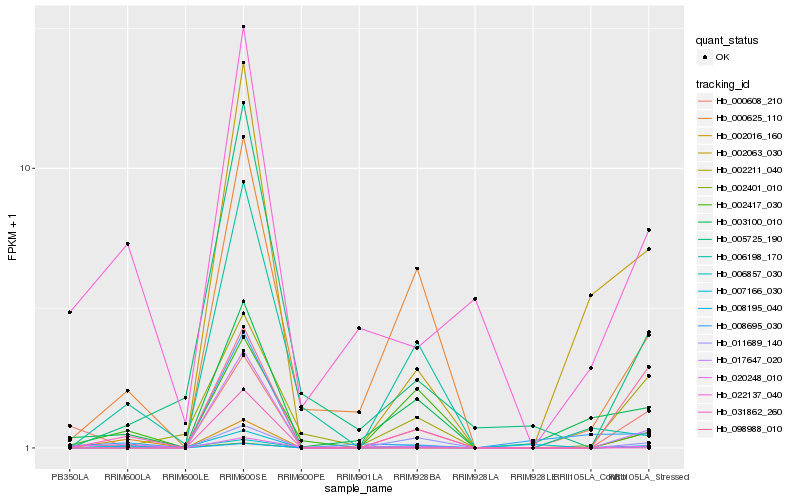
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008195_040 |
0.0 |
- |
- |
hypothetical protein VITISV_033788 [Vitis vinifera] |
| 2 |
Hb_002016_160 |
0.2405663504 |
- |
- |
- |
| 3 |
Hb_000625_110 |
0.2590510218 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005725_190 |
0.2873505569 |
- |
- |
hypothetical protein JCGZ_15992 [Jatropha curcas] |
| 5 |
Hb_006857_030 |
0.2880726612 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_002417_030 |
0.2897800463 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 7 |
Hb_002401_010 |
0.2922664362 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 8 |
Hb_002211_040 |
0.2939235314 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 9 |
Hb_003100_010 |
0.2957645375 |
- |
- |
PREDICTED: uncharacterized protein LOC101217345 [Cucumis sativus] |
| 10 |
Hb_017647_020 |
0.2986864727 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 11 |
Hb_022137_040 |
0.2993421943 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 12 |
Hb_002063_030 |
0.3048920428 |
- |
- |
- |
| 13 |
Hb_011689_140 |
0.3066983291 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 4 [Jatropha curcas] |
| 14 |
Hb_007166_030 |
0.3095044306 |
- |
- |
- |
| 15 |
Hb_000608_210 |
0.3194955698 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein FASCIATED EAR2 [Gossypium raimondii] |
| 16 |
Hb_006198_170 |
0.322701058 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070-like [Jatropha curcas] |
| 17 |
Hb_031862_260 |
0.3234006001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_098988_010 |
0.324015551 |
- |
- |
hypothetical protein JCGZ_18539 [Jatropha curcas] |
| 19 |
Hb_008695_030 |
0.3280047646 |
- |
- |
hypothetical protein JCGZ_12043 [Jatropha curcas] |
| 20 |
Hb_020248_010 |
0.3301910608 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |