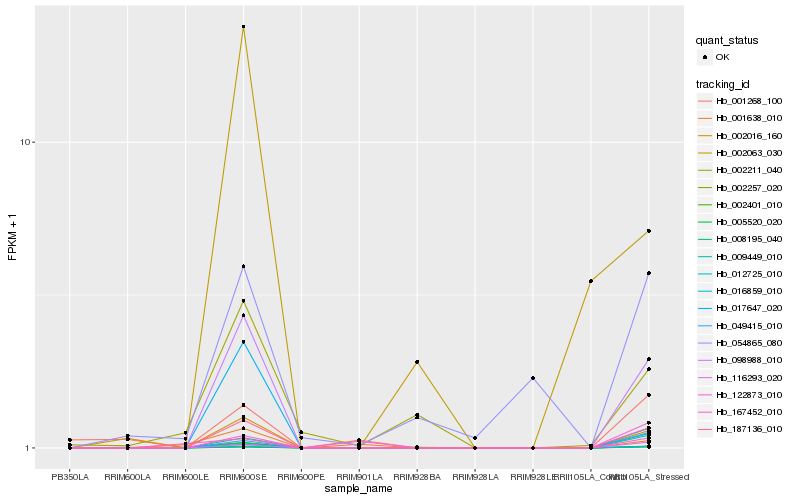
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_187136_010 |
0.0 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 2 |
Hb_098988_010 |
0.049527307 |
- |
- |
hypothetical protein JCGZ_18539 [Jatropha curcas] |
| 3 |
Hb_016859_010 |
0.0517328773 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_009449_010 |
0.1498919886 |
- |
- |
Serine/threonine-protein kinase HT1 [Gossypium arboreum] |
| 5 |
Hb_116293_020 |
0.1950762198 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 6 |
Hb_005520_020 |
0.1961812428 |
- |
- |
hypothetical protein EUGRSUZ_G03336 [Eucalyptus grandis] |
| 7 |
Hb_049415_010 |
0.229689593 |
- |
- |
- |
| 8 |
Hb_017647_020 |
0.2940998827 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 9 |
Hb_002016_160 |
0.2946917526 |
- |
- |
- |
| 10 |
Hb_167452_010 |
0.319855598 |
- |
- |
hypothetical protein JCGZ_03682 [Jatropha curcas] |
| 11 |
Hb_054865_080 |
0.3215158045 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 12 |
Hb_002257_020 |
0.3221171783 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 13 |
Hb_012725_010 |
0.3234532981 |
- |
- |
- |
| 14 |
Hb_002211_040 |
0.3258099137 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 15 |
Hb_002401_010 |
0.3303874158 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 16 |
Hb_001268_100 |
0.3315779855 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 17 |
Hb_002063_030 |
0.3416850429 |
- |
- |
- |
| 18 |
Hb_008195_040 |
0.348283672 |
- |
- |
hypothetical protein VITISV_033788 [Vitis vinifera] |
| 19 |
Hb_001638_010 |
0.349926311 |
- |
- |
- |
| 20 |
Hb_122873_010 |
0.352407629 |
- |
- |
- |