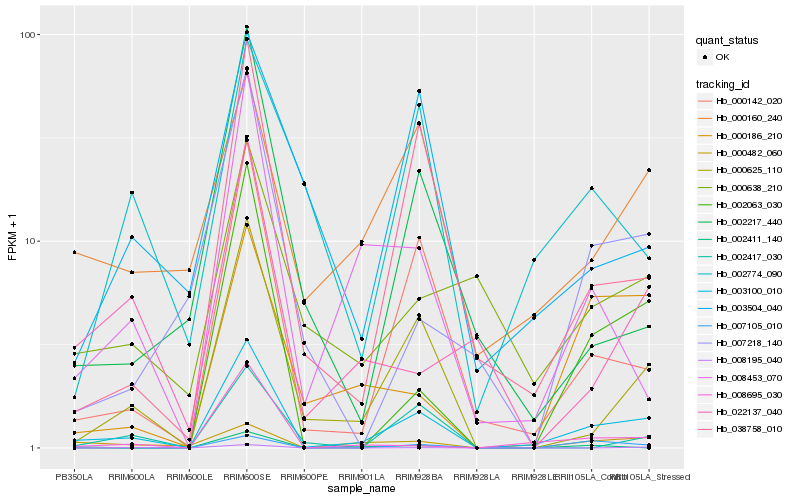
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003100_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101217345 [Cucumis sativus] |
| 2 |
Hb_000142_020 |
0.2011045573 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000625_110 |
0.2054486773 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 4 |
Hb_038758_010 |
0.2303411736 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_008695_030 |
0.2507234361 |
- |
- |
hypothetical protein JCGZ_12043 [Jatropha curcas] |
| 6 |
Hb_008453_070 |
0.2600838143 |
- |
- |
putative aquaporin [Vitis vinifera] |
| 7 |
Hb_002063_030 |
0.2690803104 |
- |
- |
- |
| 8 |
Hb_000160_240 |
0.2708786881 |
- |
- |
PREDICTED: uncharacterized protein At1g66480 [Jatropha curcas] |
| 9 |
Hb_000482_060 |
0.2738725563 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_07664 [Jatropha curcas] |
| 10 |
Hb_000186_210 |
0.2824099116 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 11 |
Hb_002217_440 |
0.2843295864 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_007218_140 |
0.2869677001 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 13 |
Hb_002417_030 |
0.2882817351 |
- |
- |
PREDICTED: purine permease 3-like [Jatropha curcas] |
| 14 |
Hb_000638_210 |
0.2914630997 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_022137_040 |
0.2926353065 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 16 |
Hb_002411_140 |
0.2944973461 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Jatropha curcas] |
| 17 |
Hb_008195_040 |
0.2957645375 |
- |
- |
hypothetical protein VITISV_033788 [Vitis vinifera] |
| 18 |
Hb_003504_040 |
0.3000591879 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 2-like [Jatropha curcas] |
| 19 |
Hb_002774_090 |
0.3016072067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007105_010 |
0.301677763 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |