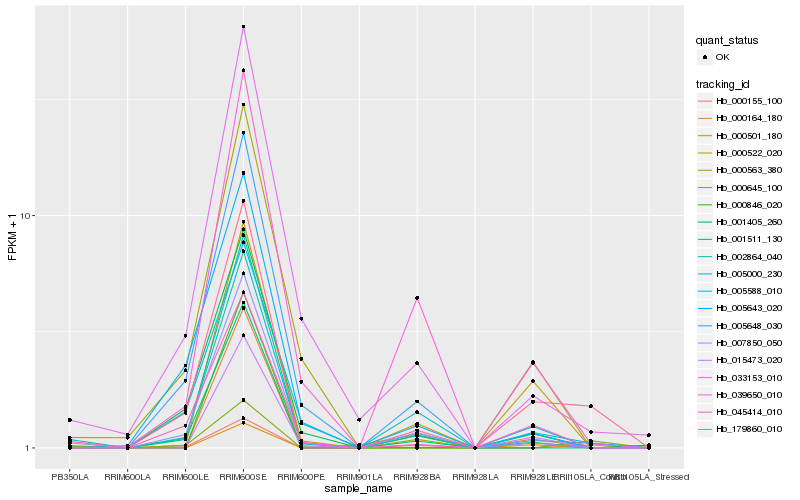
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_179860_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005000_230 |
0.1719669224 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 3 |
Hb_002864_040 |
0.1949214573 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 4 |
Hb_045414_010 |
0.1974524152 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000155_100 |
0.2017970317 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 6 |
Hb_000645_100 |
0.2033344295 |
- |
- |
enzyme inhibitor, putative [Ricinus communis] |
| 7 |
Hb_005643_020 |
0.2079036814 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 8 |
Hb_007850_050 |
0.2082137064 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 9 |
Hb_015473_020 |
0.2096117588 |
- |
- |
hypothetical protein B456_001G090400 [Gossypium raimondii] |
| 10 |
Hb_000501_180 |
0.2124106595 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 11 |
Hb_005648_030 |
0.212483044 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH18-like [Jatropha curcas] |
| 12 |
Hb_000522_020 |
0.2133082108 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 13 |
Hb_005588_010 |
0.214980441 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 14 |
Hb_001405_260 |
0.2161989862 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 15 |
Hb_001511_130 |
0.2165743307 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 16 |
Hb_039650_010 |
0.2211176566 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 17 |
Hb_000563_380 |
0.2220536271 |
- |
- |
hypothetical protein CICLE_v10007206mg [Citrus clementina] |
| 18 |
Hb_000164_180 |
0.2237690053 |
- |
- |
(+)-delta-cadinene synthase isozyme A, putative [Ricinus communis] |
| 19 |
Hb_033153_010 |
0.2243756938 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 20 |
Hb_000846_020 |
0.22492018 |
- |
- |
cytochrome P450, putative [Ricinus communis] |