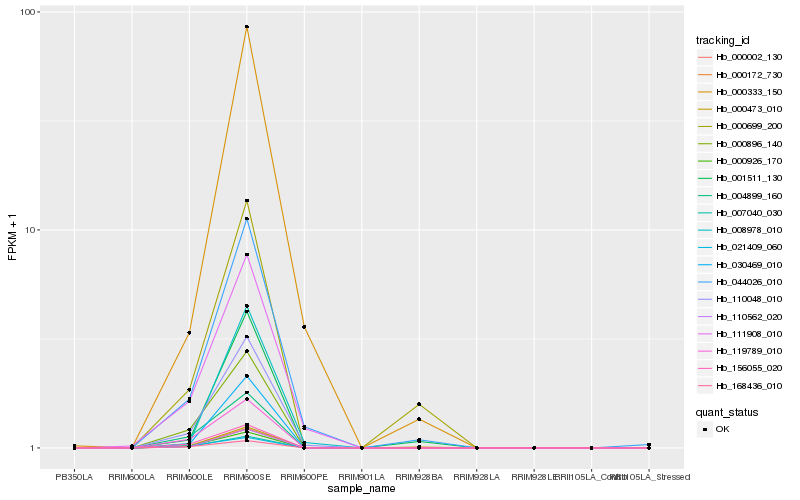
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_110562_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 2 |
Hb_000172_730 |
0.0159578856 |
- |
- |
PREDICTED: cytochrome P450 CYP736A12-like [Nicotiana sylvestris] |
| 3 |
Hb_000002_130 |
0.0295767441 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 4 |
Hb_030469_010 |
0.0431037775 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 5 |
Hb_000926_170 |
0.0723750543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007040_030 |
0.078877202 |
- |
- |
PREDICTED: uncharacterized protein LOC103415345 [Malus domestica] |
| 7 |
Hb_000896_140 |
0.0794836734 |
- |
- |
PREDICTED: zinc finger Ran-binding domain-containing protein 2 [Jatropha curcas] |
| 8 |
Hb_110048_010 |
0.0811460237 |
- |
- |
PREDICTED: uncharacterized protein LOC104887199 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_119789_010 |
0.1008531704 |
- |
- |
PREDICTED: uncharacterized protein LOC105134438 [Populus euphratica] |
| 10 |
Hb_156055_020 |
0.1095981161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004899_160 |
0.1171975222 |
- |
- |
- |
| 12 |
Hb_001511_130 |
0.1203645607 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 13 |
Hb_044026_010 |
0.130617079 |
- |
- |
PREDICTED: cytochrome P450 93A3-like [Jatropha curcas] |
| 14 |
Hb_008978_010 |
0.1365602856 |
- |
- |
transmembrane receptor, putative [Ricinus communis] |
| 15 |
Hb_021409_060 |
0.1376695343 |
- |
- |
PREDICTED: aluminum-activated malate transporter 10-like [Jatropha curcas] |
| 16 |
Hb_111908_010 |
0.138676678 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000473_010 |
0.1403006764 |
- |
- |
PREDICTED: galactolipase DONGLE, chloroplastic [Jatropha curcas] |
| 18 |
Hb_168436_010 |
0.1422731787 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000-like [Citrus sinensis] |
| 19 |
Hb_000333_150 |
0.1427186058 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 20 |
Hb_000699_200 |
0.1458369766 |
- |
- |
hypothetical protein POPTR_0019s02340g [Populus trichocarpa] |