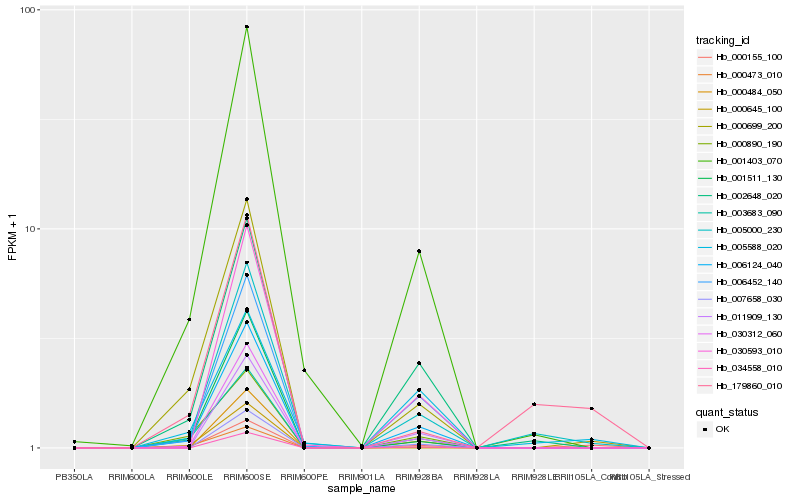
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000155_100 |
0.0 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 2 |
Hb_000645_100 |
0.1934706082 |
- |
- |
enzyme inhibitor, putative [Ricinus communis] |
| 3 |
Hb_006124_040 |
0.1940084999 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_005000_230 |
0.1985983937 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 5 |
Hb_179860_010 |
0.2017970317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_030312_060 |
0.2071446925 |
- |
- |
PREDICTED: uncharacterized protein LOC105634226 [Jatropha curcas] |
| 7 |
Hb_006452_140 |
0.2107056319 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 8 |
Hb_003683_090 |
0.2109967567 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 9 |
Hb_002648_020 |
0.2121047297 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Populus euphratica] |
| 10 |
Hb_001403_070 |
0.2130740632 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 11 |
Hb_000699_200 |
0.2139199718 |
- |
- |
hypothetical protein POPTR_0019s02340g [Populus trichocarpa] |
| 12 |
Hb_005588_020 |
0.2167861724 |
- |
- |
PREDICTED: patatin-like protein 2 [Vitis vinifera] |
| 13 |
Hb_001511_130 |
0.2216478677 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 14 |
Hb_007658_030 |
0.224148255 |
- |
- |
PREDICTED: lupeol synthase-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000484_050 |
0.2242278153 |
- |
- |
hypothetical protein CICLE_v10028549mg [Citrus clementina] |
| 16 |
Hb_000890_190 |
0.2247892374 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 17 |
Hb_011909_130 |
0.2253854542 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 18 |
Hb_030593_010 |
0.227439306 |
- |
- |
hypothetical protein JCGZ_06063 [Jatropha curcas] |
| 19 |
Hb_034558_010 |
0.2275012223 |
- |
- |
- |
| 20 |
Hb_000473_010 |
0.2277951516 |
- |
- |
PREDICTED: galactolipase DONGLE, chloroplastic [Jatropha curcas] |