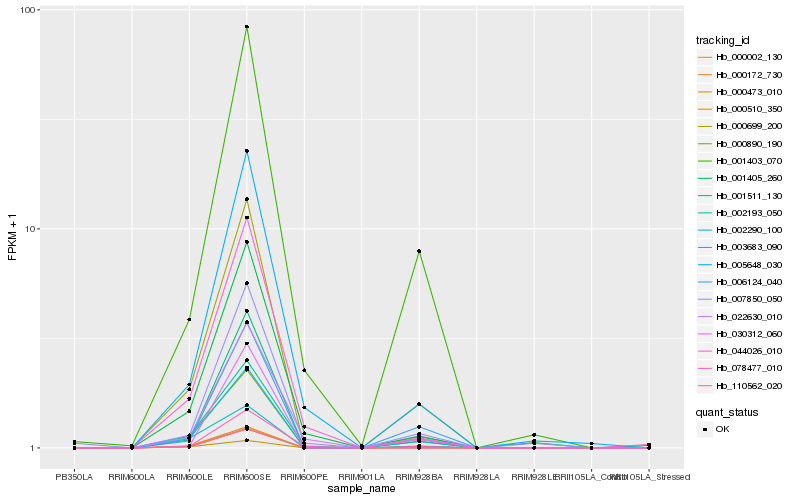
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000699_200 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s02340g [Populus trichocarpa] |
| 2 |
Hb_000473_010 |
0.031310579 |
- |
- |
PREDICTED: galactolipase DONGLE, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003683_090 |
0.0507610186 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_002193_050 |
0.0810270426 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 5 |
Hb_000890_190 |
0.0865290596 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 6 |
Hb_001511_130 |
0.0906404794 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 7 |
Hb_006124_040 |
0.1007192794 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_001403_070 |
0.1227377522 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 9 |
Hb_030312_060 |
0.1289400993 |
- |
- |
PREDICTED: uncharacterized protein LOC105634226 [Jatropha curcas] |
| 10 |
Hb_005648_030 |
0.1299602612 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH18-like [Jatropha curcas] |
| 11 |
Hb_002290_100 |
0.1312700199 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 12 |
Hb_022630_010 |
0.1348393363 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001405_260 |
0.1386218119 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 14 |
Hb_078477_010 |
0.1395545501 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 15 |
Hb_007850_050 |
0.1409620173 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 16 |
Hb_110562_020 |
0.1458369766 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 17 |
Hb_000002_130 |
0.1461633225 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 18 |
Hb_000510_350 |
0.1473084811 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 19 |
Hb_044026_010 |
0.1476429817 |
- |
- |
PREDICTED: cytochrome P450 93A3-like [Jatropha curcas] |
| 20 |
Hb_000172_730 |
0.1480813498 |
- |
- |
PREDICTED: cytochrome P450 CYP736A12-like [Nicotiana sylvestris] |