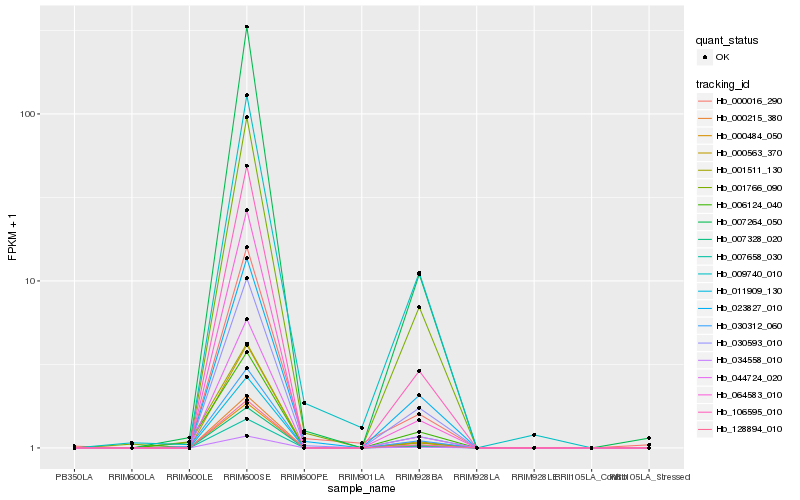
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030312_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634226 [Jatropha curcas] |
| 2 |
Hb_034558_010 |
0.0653849297 |
- |
- |
- |
| 3 |
Hb_128894_010 |
0.0655254555 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_106595_010 |
0.0691762736 |
- |
- |
hypothetical protein B456_003G065300 [Gossypium raimondii] |
| 5 |
Hb_044724_020 |
0.0740568086 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_007264_050 |
0.075790538 |
- |
- |
- |
| 7 |
Hb_001766_090 |
0.0773430745 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 8 |
Hb_007658_030 |
0.0795959291 |
- |
- |
PREDICTED: lupeol synthase-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000215_380 |
0.0799318469 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 10 |
Hb_006124_040 |
0.0802322262 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_007328_020 |
0.0807230548 |
- |
- |
- |
| 12 |
Hb_000563_370 |
0.0811921476 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |
| 13 |
Hb_000484_050 |
0.0840134397 |
- |
- |
hypothetical protein CICLE_v10028549mg [Citrus clementina] |
| 14 |
Hb_030593_010 |
0.0865460165 |
- |
- |
hypothetical protein JCGZ_06063 [Jatropha curcas] |
| 15 |
Hb_001511_130 |
0.0870770849 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 16 |
Hb_011909_130 |
0.0963073769 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 17 |
Hb_064583_010 |
0.099410808 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 7 [Vitis vinifera] |
| 18 |
Hb_023827_010 |
0.1001465094 |
- |
- |
Cucumber peeling cupredoxin, putative [Ricinus communis] |
| 19 |
Hb_009740_010 |
0.1017689285 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 20 |
Hb_000016_290 |
0.1069112872 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |