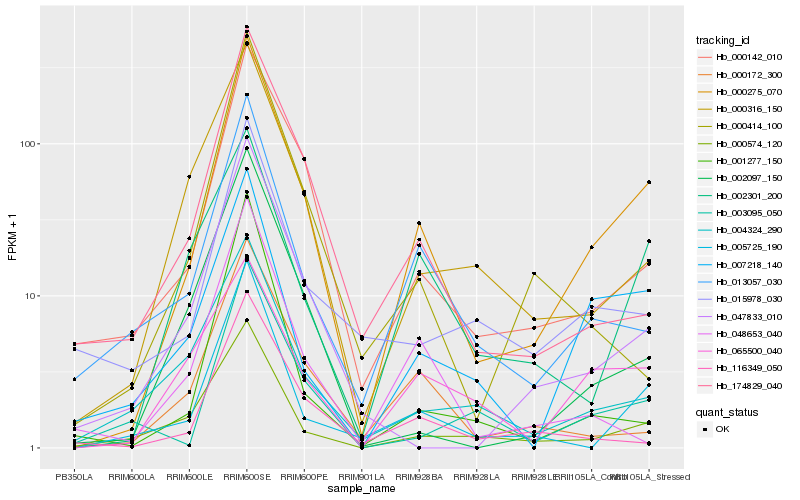
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000275_070 |
0.0 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 2 |
Hb_005725_190 |
0.1717937184 |
- |
- |
hypothetical protein JCGZ_15992 [Jatropha curcas] |
| 3 |
Hb_000142_010 |
0.1761089684 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 4 |
Hb_000574_120 |
0.1908666936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_013057_030 |
0.1910560744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007218_140 |
0.191347317 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 7 |
Hb_000172_300 |
0.2009591251 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 8 |
Hb_174829_040 |
0.2020796252 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 9 |
Hb_047833_010 |
0.2036081871 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 10 |
Hb_002301_200 |
0.2052628779 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 11 |
Hb_003095_050 |
0.2055302015 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 12 |
Hb_002097_150 |
0.2059266366 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 13 |
Hb_116349_050 |
0.2066935081 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_000316_150 |
0.2068326932 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_015978_030 |
0.2122067539 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 16 |
Hb_004324_290 |
0.2168490018 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 17 |
Hb_001277_150 |
0.221240156 |
- |
- |
hypothetical protein VITISV_043492 [Vitis vinifera] |
| 18 |
Hb_065500_040 |
0.2215206933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_048653_040 |
0.2227370987 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 20 |
Hb_000414_100 |
0.2230573768 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |