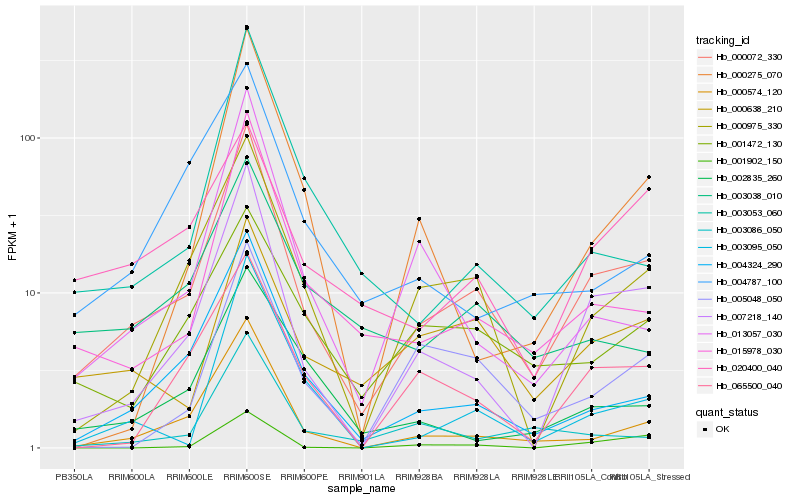
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_330 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007218_140 |
0.1410019843 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 3 |
Hb_000574_120 |
0.1533959439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000975_330 |
0.1659857125 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT12 [Jatropha curcas] |
| 5 |
Hb_004324_290 |
0.1667471943 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 6 |
Hb_015978_030 |
0.1700037997 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 7 |
Hb_065500_040 |
0.1921154739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000638_210 |
0.1988402972 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_001472_130 |
0.2106696391 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 10 |
Hb_003086_050 |
0.2122761139 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_013057_030 |
0.2140288153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003095_050 |
0.2141243389 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 13 |
Hb_005048_050 |
0.2153396721 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 14 |
Hb_003053_060 |
0.2155605805 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 15 |
Hb_003038_010 |
0.2178833174 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_002835_260 |
0.2179462669 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 17 |
Hb_020400_040 |
0.2186568116 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 18 |
Hb_004787_100 |
0.2239718876 |
- |
- |
PREDICTED: transcription factor bHLH47 [Jatropha curcas] |
| 19 |
Hb_000275_070 |
0.227612008 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 20 |
Hb_001902_150 |
0.2314977641 |
- |
- |
PREDICTED: cytokinin hydroxylase-like [Jatropha curcas] |