| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
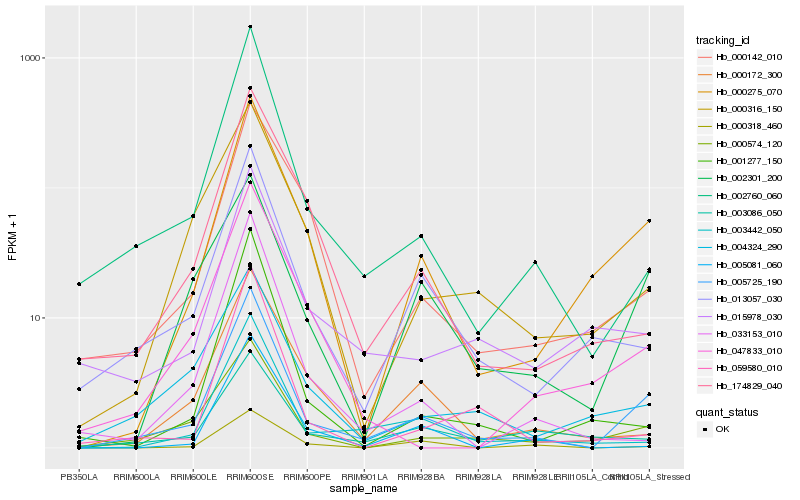
Hb_005725_190 |
0.0 |
- |
- |
hypothetical protein JCGZ_15992 [Jatropha curcas] |
| 2 |
Hb_000275_070 |
0.1717937184 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 3 |
Hb_000574_120 |
0.1739181023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002760_060 |
0.1740643526 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 5 |
Hb_003442_050 |
0.1934237096 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 6 |
Hb_013057_030 |
0.1951999576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002301_200 |
0.2072168674 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 8 |
Hb_004324_290 |
0.2158318585 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF4 [Jatropha curcas] |
| 9 |
Hb_000172_300 |
0.2165467259 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 10 |
Hb_001277_150 |
0.2178203967 |
- |
- |
hypothetical protein VITISV_043492 [Vitis vinifera] |
| 11 |
Hb_015978_030 |
0.2189619807 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 12 |
Hb_000142_010 |
0.219467458 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 13 |
Hb_000316_150 |
0.2200617703 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_033153_010 |
0.2207823334 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 15 |
Hb_174829_040 |
0.2213879731 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 16 |
Hb_059580_010 |
0.2215564399 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 17 |
Hb_005081_060 |
0.2258140887 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 18 |
Hb_047833_010 |
0.2261350186 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 19 |
Hb_000318_460 |
0.2263614239 |
- |
- |
hypothetical protein CICLE_v10028333mg [Citrus clementina] |
| 20 |
Hb_003086_050 |
0.2270899526 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |