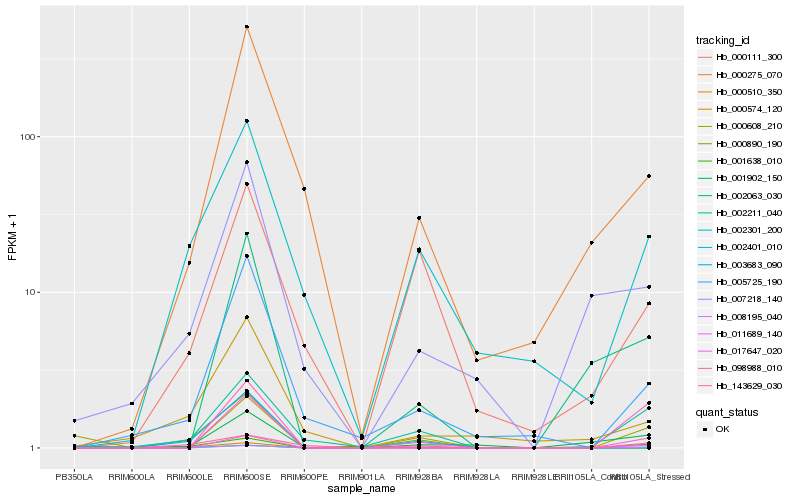
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002401_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 2 |
Hb_002211_040 |
0.1962636876 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 3 |
Hb_002301_200 |
0.2185120211 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 4 |
Hb_143629_030 |
0.2321145125 |
- |
- |
hypothetical protein JCGZ_23143 [Jatropha curcas] |
| 5 |
Hb_005725_190 |
0.2615937334 |
- |
- |
hypothetical protein JCGZ_15992 [Jatropha curcas] |
| 6 |
Hb_000111_300 |
0.2812091446 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 7 |
Hb_001902_150 |
0.2849264763 |
- |
- |
PREDICTED: cytokinin hydroxylase-like [Jatropha curcas] |
| 8 |
Hb_011689_140 |
0.2880342499 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 4 [Jatropha curcas] |
| 9 |
Hb_008195_040 |
0.2922664362 |
- |
- |
hypothetical protein VITISV_033788 [Vitis vinifera] |
| 10 |
Hb_000275_070 |
0.2941019863 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 11 |
Hb_007218_140 |
0.2999845143 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 12 |
Hb_001638_010 |
0.3023021737 |
- |
- |
- |
| 13 |
Hb_000574_120 |
0.3036728176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002063_030 |
0.3042847747 |
- |
- |
- |
| 15 |
Hb_000608_210 |
0.305124591 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein FASCIATED EAR2 [Gossypium raimondii] |
| 16 |
Hb_017647_020 |
0.3074742172 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 17 |
Hb_098988_010 |
0.30917204 |
- |
- |
hypothetical protein JCGZ_18539 [Jatropha curcas] |
| 18 |
Hb_000890_190 |
0.3185282708 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 19 |
Hb_000510_350 |
0.3202671193 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 20 |
Hb_003683_090 |
0.323220352 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |