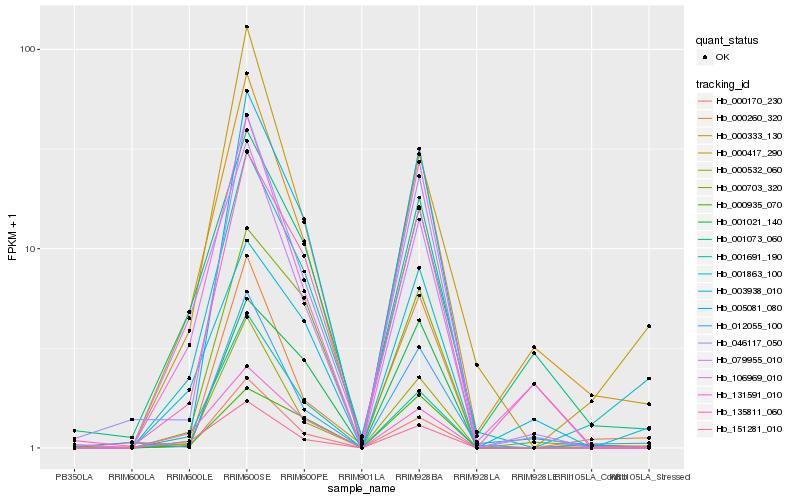
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000333_130 |
0.1409122574 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_003938_010 |
0.1551654016 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 4 |
Hb_012055_100 |
0.1616420879 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 5 |
Hb_005081_080 |
0.1618555131 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 6 |
Hb_000703_320 |
0.1636612996 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_000260_320 |
0.1638867587 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 8 |
Hb_000417_290 |
0.1652951726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_135811_060 |
0.1668919099 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 10 |
Hb_001073_060 |
0.1673534003 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 11 |
Hb_151281_010 |
0.1692978915 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 12 |
Hb_131591_010 |
0.1702779675 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 13 |
Hb_000532_060 |
0.170848864 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 14 |
Hb_000170_230 |
0.1717781427 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 15 |
Hb_046117_050 |
0.1726744724 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 16 |
Hb_001021_140 |
0.1743626863 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 17 |
Hb_001691_190 |
0.1768291529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000935_070 |
0.1783809117 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 19 |
Hb_079955_010 |
0.1792542116 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 20 |
Hb_106969_010 |
0.1793973287 |
- |
- |
Cytochrome P450 76C4 [Glycine soja] |