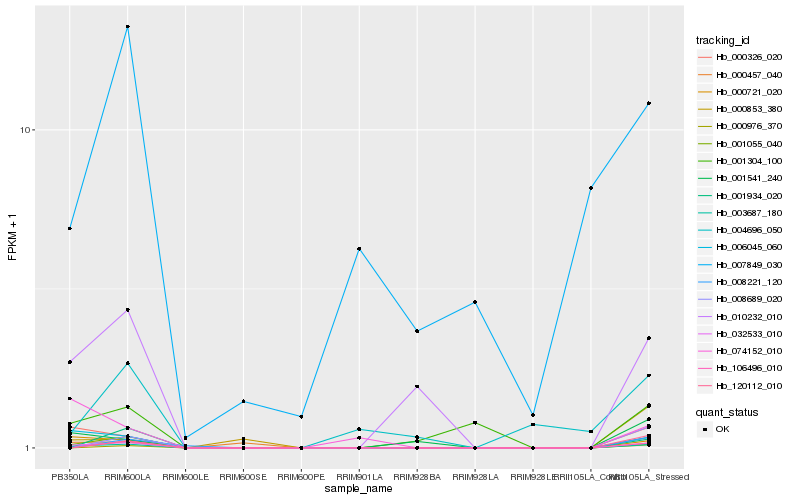
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032533_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 2 |
Hb_003687_180 |
0.0114297289 |
- |
- |
hypothetical protein JCGZ_02335 [Jatropha curcas] |
| 3 |
Hb_000721_020 |
0.2034688786 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 4 |
Hb_006045_060 |
0.2123573063 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |
| 5 |
Hb_000326_020 |
0.2156117239 |
- |
- |
- |
| 6 |
Hb_001055_040 |
0.2158664958 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 7 |
Hb_010232_010 |
0.2640887113 |
- |
- |
- |
| 8 |
Hb_001541_240 |
0.3387100819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_106496_010 |
0.339364387 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 10 |
Hb_000457_040 |
0.339580286 |
- |
- |
- |
| 11 |
Hb_001304_100 |
0.3456790313 |
- |
- |
- |
| 12 |
Hb_000976_370 |
0.3630093589 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 13 |
Hb_120112_010 |
0.3644856003 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_008689_020 |
0.3657139912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001934_020 |
0.3659728799 |
- |
- |
PREDICTED: uncharacterized protein LOC105178413 [Sesamum indicum] |
| 16 |
Hb_074152_010 |
0.3772181909 |
- |
- |
- |
| 17 |
Hb_008221_120 |
0.4011946158 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g63170-like [Jatropha curcas] |
| 18 |
Hb_000853_380 |
0.4042502237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004696_050 |
0.4056845159 |
- |
- |
- |
| 20 |
Hb_007849_030 |
0.4105426445 |
- |
- |
l-allo-threonine aldolase, putative [Ricinus communis] |